Achintya Mahanti, Siddhartha Narayan Joardar, Samiran Bandyopadhyay, Jaydeep Banerjee, Sarbaswarup Ghosh, Tapan Kumar Dutta, Indranil Samanta
ABSTRACT. The transmission of zoonotic bacteria through consumption of raw milk is complicated by the dissemination of antimicrobial-resistant bacteria. The present study was conducted to detect the occurrence of antimicrobial-resistant bacteria (ESBL-/AmpC-producing Klebsiella spp.) in cow’s milk originating from healthy or infected (mastitis) cattle in India. In total, 450 milk samples were collected from apparently healthy cattle and cattle suffering from clinical or sub-clinical mastitis. Out of 455 Klebsiella spp., 67 (14.73%) isolates were found to be ESBL producers in the double-disc diffusion test. The occurrence of ESBL-producing Klebsiella spp. was significantly (p < 0.05) higher in milk samples collected from cattle suffering with mastitis than in healthy cattle. Among the ESBL-producing Klebsiella spp., 56 (83.6%) isolates were also detected that produced AmpC β-lactamases. All the ESBL and AmpC-producing Klebsiella spp. possessed blaCTX-M (100%) and blaAmpC (100%), respectively. The present study revealed a higher occurrence of class 1 integron in ESBL-producing Klebsiella spp. isolates. All ESBL-producing- Klebsiella spp. isolates were multi-drug resistant. The ciprofloxacin- and/or levofloxacin-resistant Klebsiella spp. isolates possessed the quinolone resistance gene (qnrS). The co-trimoxazole-resistant isolates possessed the sul1 and sul2 genes. Phylogenetic analysis of the studied isolates revealed that strains isolated from the same location had a clonal relationship. The study increases consumer awareness of the need to avoid raw milk consumption to prevent the spread of antimicrobial resistance in the community.
Keywords: antimicrobial resistance; ESBL; Klebsiella; MIC; raw milk.
Cite
ALSE and ACS Style
Mahanti, A.; Joardar, S.N.; Bandyopadhyay, S.; Banerjee, J.; Ghosh, S.; Dutta, T.K.; Samanta, I. Raw Bovine Milk as a Reservoir of Multi-Drug Resistant, Beta-Lactamase-Producing Klebsiella. Journal of Applied Life Sciences and Environment 2024, 57 (1), 19-36.
https://doi.org/10.46909/alse-571122
AMA Style
Mahanti A, Joardar SN, Bandyopadhyay S, Banerjee J, Ghosh S, Dutta TK, Samanta I. Raw Bovine Milk as a Reservoir of Multi-Drug Resistant, Beta-Lactamase-Producing Klebsiella. Journal of Applied Life Sciences and Environment. 2024; 57 (1): 19-36.
https://doi.org/10.46909/alse-571122
Chicago/Turabian Style
Mahanti, Achintya, Siddhartha Narayan Joardar, Samiran Bandyopadhyay, Jaydeep Banerjee, Sarbaswarup Ghosh, Tapan Kumar Dutta, and Indranil Samanta. 2024. “Raw Bovine Milk as a Reservoir of Multi-Drug Resistant, Beta-Lactamase-Producing Klebsiella” Journal of Applied Life Sciences and Environment 57, no. 1: 19-36.
https://doi.org/10.46909/alse-571122
View full article (HTML)
Raw Bovine Milk as A Reservoir of Multi-Drug Resistant, Beta-Lactamase-Producing Klebsiella
Achintya MAHANTI1, Siddhartha Narayan JOARDAR1, Samiran BANDYOPADHYAY2, Jaydeep BANERJEE2, Sarbaswarup GHOSH3, Tapan Kumar DUTTA4 and Indranil SAMANTA1*
1Department of Veterinary Microbiology, West Bengal University of Animal and Fishery Sciences, 37, K.B. Sarani, Belgachia, Kolkata-700037, West Bengal, India; email: dr.amahanti@gmail.com; joardar69@gmail.com
2Eastern Regional Station, Indian Veterinary Research Institute, Belgachia, Kolkata, West Bengal, India; email: samiranvet@gmail.com; jaydeepige@gmail.com
3Sasya Shyamala Krishi Vigyan Kendra, Ramakrishna Mission Vivekananda Educational and Research Institute, Arapanch, Sonarpur, West Bengal, India; email: dr.sarba@rediffmail.com
4Department of Veterinary Microbiology, Central Agricultural University, Selesih, Aizawl, India; email: tapandutta@rediffmail.com
*Correspondence: isamanta76@gmail.com, drisamanta@wbuafscl.ac.in
Received: Dec. 27, 2023. Revised: Jan. 28, 2024. Accepted: Jan. 30, 2024. Published online: Feb. 28, 2024
ABSTRACT. The transmission of zoonotic bacteria through consumption of raw milk is complicated by the dissemination of antimicrobial-resistant bacteria. The present study was conducted to detect the occurrence of antimicrobial-resistant bacteria (ESBL-/AmpC-producing Klebsiella spp.) in cow’s milk originating from healthy or infected (mastitis) cattle in India. In total, 450 milk samples were collected from apparently healthy cattle and cattle suffering from clinical or sub-clinical mastitis. Out of 455 Klebsiella spp., 67 (14.73%) isolates were found to be ESBL producers in the double-disc diffusion test. The occurrence of ESBL-producing Klebsiella spp. was significantly (p < 0.05) higher in milk samples collected from cattle suffering with mastitis than in healthy cattle. Among the ESBL-producing Klebsiella spp., 56 (83.6%) isolates were also detected that produced AmpC β-lactamases. All the ESBL and AmpC-producing Klebsiella spp. possessed blaCTX-M (100%) and blaAmpC (100%), respectively. The present study revealed a higher occurrence of class 1 integron in ESBL-producing Klebsiella spp. isolates. All ESBL-producing- Klebsiella spp. isolates were multi-drug resistant. The ciprofloxacin- and/or levofloxacin-resistant Klebsiella spp. isolates possessed the quinolone resistance gene (qnrS). The co-trimoxazole-resistant isolates possessed the sul1 and sul2 genes. Phylogenetic analysis of the studied isolates revealed that strains isolated from the same location had a clonal relationship. The study increases consumer awareness of the need to avoid raw milk consumption to prevent the spread of antimicrobial resistance in the community.
Keywords: antimicrobial resistance; ESBL; Klebsiella; MIC; raw milk.
INTRODUCTION
Raw bovine milk as a reservoir of multi-drug resistant, beta-lactamase-producing Klebsiella
Food items such as milk function as ideal media for bacterial growth due to the presence of nutrients in optimum proportions. The microbes in raw milk originate from the bovine udder during intra-mammary infection (IMI) or diverse external sources such as air, contaminated hands of milkers, milking equipment, bulk tanks etc. The poor animal husbandry conditions associated with contaminated feed and drinking water, as well as the hind quarter hygiene of milch animals, play a significant role in the development of IMI, as environmental pathogens can enter the udder through the teat canal (Parekh and Subhash, 2008). The transmission of zoonotic bacteria through the consumption of raw milk and/or milk products is complicated by the dissemination of mobile genetic elements carrying antimicrobial resistance genes. The therapeutic efficacy and short withdrawal period of third- and fourth-generation cephalosporin β-lactam antibiotics make antimicrobial resistance a critical factor in dairy farming, especially during the treatment of mastitis, the most significant multi-etiological intramammary infection compromising the microbiological quality and quantity of produced milk. Overuse or misuse of antibiotics produces selection pressure that can generate commensal Enterobacteriaceae, including Klebsiella pneumoniae, that produce extended-spectrum β-lactamase (ESBL) or aminopenicillin-inactivating cephalosporinase (AmpC) (Liebana et al., 2013). Data on the occurrence of ESBL-/AmpC-producing Enterobacteriaceae, particularly Klebsiella pneumoniae, in raw cattle milk are inconsistent (Gundogan and Yakar, 2007). Most of the studies published thus far have focused on milk samples collected from cattle suffering with mastitis, so their results cannot be extrapolated to apparently normal milk entering the food chain, collected from healthy cattle reared in the same geographical location (Dahmen et al., 2013; Ohnishi et al., 2013).
In the Enterobacteriaceae group of bacteria, including Klebsiella spp., Escherichia coli and Salmonella spp., extended-spectrum beta-lactamases (ESBLs) are major antimicrobial resistance determinants which are transmitted through horizontal gene transfer. The ESBL enzyme can produce resistance to a variety of β-lactam antibiotics, such as penicillins, higher-generation cephalosporins and aztreonam. Classical ESBL varieties include TEM (except TEM-1), SHV (except SHV-1 and 2) and CTX-M (EFSA, 2011). AmpC β-lactamase-producing organisms (ACBL) can generate resistance against β-lactamase inhibitors, such as clavulanic acid, in addition to cephalosporins, penicillins, cephamycins and monobactams. Overexpression of the chromosomal AmpC gene (blaAmpC) or possession of plasmid-mediated (CITM or blaCMY-2) genes are found associated with the generation of resistance by ACBL producers (Schmid et al., 2013). The emergence and spread of carbapenem-resistant Enterobacteriaceae (CRE) is a cause of serious clinical and public health concerns, as carbapenem is considered as a last-resort therapeutic option against ESBL-producing bacteria. Carbapenem resistance in Klebsiella spp. is associated with the production of enzymes such as metallo-β-lactamase and K. pneumoniae carbapenemase (KPC) (Birgy et al., 2012).
Moreover, possession of ESBL determinants in bacterial plasmids is often associated with resistance to unrelated classes of antibiotics, for example, fluoroquinolones, aminoglycosides and sulfmethoxazole-trimethoprim (Coque et al., 2008). Mobile genetic elements, such as class 1 integrons, aid in the transmission of ESBL-/ACBL- producing organisms (EFSA, 2011).
The Indian dairy sector is mostly unorganized, and farmers are the major stakeholders. The ownership of milch animals (cattle and water buffalo) is fragmented, and large numbers of farmers rear only a few animals (0.6–2.0 animals per holding) for milking and draught purposes (Landes et al., 2017). About 35% of the milk produced is collected by local co-operatives for processing and distribution to consumers, whereas the majority of the milk (40%) produced is consumed within the farmer’s household or distributed locally without processing. During milk processing in dairy plants, the majority of bacteria, including Enterobacteriaceae, are destroyed at pasteurization temperature. The consumption of raw milk is still a traditional practice in Indian villages, and this practice has increased recently among urban youth, who believe in the beneficial health effects of raw milk. As the local food animals were found to harbour ESBL-/ACBL-producing Enterobacteriaceae (Bandyopadhyay et al., 2015; Kar et al., 2015; Samanta et al., 2015 ), animal products such as milk may constitute an important reservoir of ESBL-/AmpC-producing Enterobacteriaceae.
The present study was conducted to detect the occurrence of antimicrobial-resistant bacteria (ESBL-/AmpC-producing Klebsiella spp.) in cow’s milk originating from healthy or infected (mastitis) cattle in all agro-climatic zones of West Bengal, one of the major milk-producing states in India (Landes et al., 2017).
The study also intended to reveal the occurrence of integron, production of carbapenemase and metallo-beta-lactamase enzymes, MIC of antimicrobials and phylogenetic relationships among the Klebsiella spp. isolates.
MATERIALS AND METHODS
Sampling
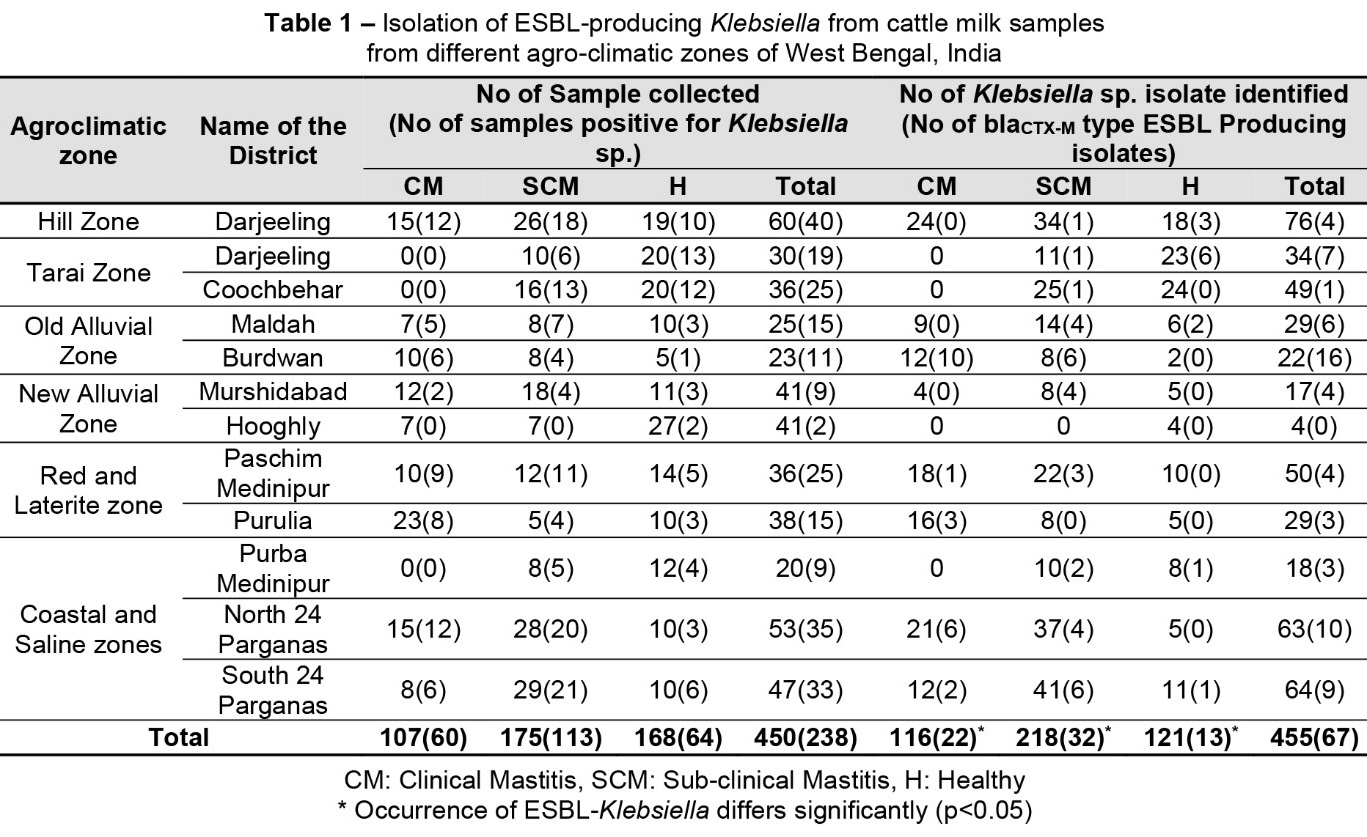
In the study, 450 milk samples were collected from apparently healthy (n = 168, without any visible clinical signs) cattle and infected cattle with clinical (n = 107) or sub-clinical mastitis (n = 175) from six agro-climatic zones (North 24 Parganas, Darjeeling, Coochbehar, Maldah, Mursidabad, Bardhaman, Paschim Medinipur, Purulia, Purba Medinipur, South 24 Parganas districts) of West Bengal (India), irrespective of the age and breed of the animals, during 2016 (Table 1). The institutional animal ethics committee approved the study (WBUAFS/IAEC/032/2013-14, dated 22/10/2013). After discarding of the first two strips, the milk samples were collected directly into sterile vials. Sub-clinical mastitis was detected by using the California mastitis test (MAST INDEX, Tulip Diagnostics Private Limited, India) following the manufacturer’s instructions.
From the clinical mastitis cases, the milk samples were collected following the guidelines of the National Mastitis Council (NMC, 1990). Cattle without visible clinical signs and with a negative CMT were considered as ‘apparently healthy’. A pooled milk sample was prepared by mixing all four quarter milk samples, and the pooled sample was brought into the laboratory, maintaining the cold chain.
Most of the cattle were reared in backyard farming systems where the farmers kept 2–3 cattle per household. All the cattle were milked by hand by the farmers. In cattle suffering with clinical mastitis, mostly higher generation cephalosporins (ceftizoxime) and sometimes enrofloxacin and tetracycline were used for therapy during milk sample collection.
The data were collected from farmers during milk sample collection using the ‘drug bag’ method, where the bags containing the foils/vials of common antibiotics were displayed to the farmers to identify the antibiotics used in his/her cattle. Cattle suffering with sub-clinical mastitis were not treated, as no clinical signs were present.
Isolation, Identification and PCR-Based Confirmation of Klebsiella sp.
Klebsiella-selective agar (HiMedia, India) was used for isolation of the bacteria from the collected milk samples. More than one characteristic purple-magenta-coloured colony was selected and streaked on a nutrient agar slant (HiMedia, India) for morphological and biochemical confirmation.
Morphological confirmation was performed by Gram staining and biochemical confirmation by indole, methyl-red, Voges-Proskauer and citrate utilization tests (Quinn et al., 1994). PCR was used to confirm the isolates as Klebsiella spp. following the cycling conditions reported earlier, with some modifications (Brisse and Verhoef, 2001).
The annealing temperature was modified using the positive control (ATCC-BAA-1705) in a gradient thermal cycler (Mastercycler Nexus, Eppendorf).
Double-Disc Diffusion Test
The disc diffusion assay with cefotaxime (HiMedia, India; 30 μg,) and ceftazidime (HiMedia, India; 30 μg,) with or without clavulanate (HiMedia, India; 10 μg,) was conducted to detect ESBL production by Klebsiella spp. isolates (Patel et al., 2014).
Phenotypic detection of AmpC production was conducted by cefoxitin-cloxacillin double-disc synergy (CC-DDS) (Tan et al., 2009). PCR was used to confirm Klebsiella pneumoniae among all the ESBL-producing Klebsiella spp. isolates (Liu et al., 2008).
Detection of Beta-Lactamase (Blactx-M, Blatem, Blashv), Chromosomal Blaampc, Plasmid-Mediated Ampc Β-Lactamase (CITM) and Integron Genes in the Klebsiella spp. Isolates
PCR was conducted for detection of blaCTX-M, blaTEM and blaSHV in Klebsiella spp. isolates showing a positive reaction in the double-disc diffusion test (Cao et al., 2002; Weill et al., 2004) Standard PCR for detection of blaAmpC and CITM genes was also conducted in Klebsiella spp. isolates showing phenotypical AmpC production (Féria et al., 2002; van et al., 2008). Class I integron was detected by PCR in all the ESBL-producing Klebsiella spp. isolates (Mazel et al., 2000).
Antimicrobial Sensitivity of ESBL-Producing Klebsiella spp. Isolates
All the ESBL-producing Klebsiella spp. isolates were tested against different antimicrobials by the disc diffusion method (Patel et al., 2014). The antimicrobial agents used were cefoxitin (30 µg), co-trimoxazole (25 µg), streptomycin (10 µg), ertapenem (10 µg), ceftriaxone/tazobactam (30/10 µg), ciprofloxacin (5 µg), tetracycline (30 µg), cefoperazone (75 µg), chloramphenicol (30 µg), cefepime (30 µg), gentamicin (10 µg), amikacin (30 µg), levofloxacin (5 µg), penicillin-G (10U), ampicillin/sulbactam (10/10 µg), doxycycline hydrochloride (10 µg), tobramycin (10 µg), ceftizoxime (30 µg), amoxycillin/clavulanic acid (20/10 µg), piperacillin (100 µg) and ceftazidime (30 µg) (HiMedia, India). The criteria for susceptibility/resistance were detected following the CLSI guideline (Patel et al., 2014).
Phenotypic Detection of Carbapenemase and Metallo-Beta-Lactamase in ESBL-Producing Klebsiella sp. Isolates
All the ESBL-producing Klebsiella spp. isolates showing phenotypical resistance against ertapenem were subjected to the modified hodge test (MHT) and combination disc diffusion test (CDDT) (EDTA-750 µg + IMP-10 µg and IMP-10 µg) to confirm carbapenemase and metallo-beta-lactamase (MBL) production, respectively(Birgy et al., 2012).
Detection of PMQR (qnrA, qnrB, qnrS) and Sulphonamide Resistance Genes (sul1, sul2, sul3)
The quinolone (qnrA, qnrB, qnrS) and sulphonamide resistance genes (sul1, sul2, sul3) were detected by PCR in all the ciprofloxacin- and/or levofloxacin- and co-trimoxazole-resistant ESBL-producing Klebsiella spp. isolates (Kar et al., 2015; Frank et al., 2007).
Detection of Minimum Inhibitory Concentration of Cefotaxime, Ceftazidime, Ceftriaxone and Ampicillin
The MIC of cefotaxime, ceftazidime, ceftriaxone and ampicillin was determined against ESBL-producing Klebsiella spp. isolates using HiComb™ MIC Strip (HiMedia, India) and Ezy MIC strips (HiMedia, India) as per the guidelines of the manufacturer.
The MIC of ciprofloxacin was determined against Klebsiella spp. isolates possessing qnr genes using Ezy MIC paper strips (HiMedia, India) as per the guidelines of manufacturer.
Phylogeny of ESBL-Producing Klebsiella spp. Isolates
All ESBL-producing Klebsiella spp. isolates were typed by RAPD-PCR (Lim et al., 2009), and images were analysed by using Doc-itLs image analysis software (UVP, UK). The phylogeny of the isolates was established by comparing the differences in the RAPD banding pattern. The neighbour joining method was used to construct an unrooted phylogenetic tree.
Statistical Analysis
The occurrence of ESBL-producing Klebsiella spp. in milk samples collected from apparently healthy and infected cattle was compared by chi-square test and a descriptive analysis was performed using SPSS software (SPSS Inc.).
RESULTS AND DISCUSSION
The present study detected the occurrence of Klebsiella spp. in 238 (238/450, 52.8%) milk samples collected from healthy and diseased cattle. From the 238 samples, a total of 455 Klebsiella spp. isolates were identified phenotypically and confirmed by specific PCR. The isolation rate of Klebsiella spp. (38.09%–64.57%) varies according to the health status of the studied animals. Significant (p < 0.05) differences in the occurrence of Klebsiella spp. were detected between the three groups of cattle according to their health status (Table 1). More than one colony was selected from each composite milk sample; therefore, the isolated strains outnumbered the collected samples. The isolation rate (20%–60%) of Klebsiella spp. varied widely in raw milk samples collected from farmers, bulk tank milk and milk products (cheese, khoa) in Jordan, Sudan, Sri Lanka, Egypt and Turkey (Ahmed et al., 2016; Badri et al., 2017; El-Sukhon, 2003; Jayaweera et al., 2018; Tepeli and Zorba, 2018). The existence of Klebsiella in the bovine udder is facilitated by the lac and fec iron-enterobactin operon, which helps Klebsiella spp. to utilize milk lactose more effectively than other commensal bacteria and might explain the high occurrence of the studied bacteria in the collected milk samples (Holt et al., 2015).
Out of 455 Klebsiella sp. isolates, 67(67/455, 14.73%) were found to be phenotypical ESBL producers in the double-disc diffusion test. The frequency of ESBL-producing Enterobacteriaceae in raw milk from dairy farms varies from 0 to 9.5% throughout the world depending on husbandry conditions, hind quarter hygiene, the use of milking equipment and the use of antimicrobials on farms (Geser et al., 2012; Skočková et al., 2015; Odenthal et al., 2016; Tepeli et al., 2018). The studies with detection of ESBL-producing Klebsiella spp. in raw milk described a highly variable occurrence between 0 and 45% worldwide, which is concurrent with the findings of the present study (Badri et al., 2017; Diab et al., 2017; Gundogan and Yakar, 2007; Özpınar et al., 2017). The occurrence of ESBL-producing Klebsiella spp. was significantly (p < 0.05) higher in milk samples collected from cattle suffering with mastitis (54/334, 16.1%) than in samples from healthy ones (13/121, 10.7%, Table 1). More samples were collected from infected cattle (clinical/sub-clinical mastitis) than from apparently healthy cattle, which may be one of the reasons, other than exposure to antibiotics, why more ESBL producers were obtained from infected milk samples. However, cattle with sub-clinical mastitis did not undergo any antibiotic therapy as they had no clinical signs. Earlier studies, including some form from India, also revealed mastitis milk as a potential source of ESBL-producing organisms due to high therapeutic antimicrobial exposure of the infected animals (Bandyopadhyay et al., 2015; Locatelli et al., 2010). Earlier studies detected faecal carriage of ESBL/ACBL-producing Enterobacteriaceae by food animals in India (Kar et al., 2015; Samanta et al., 2015). Moreover, Klebsiella spp. isolated from human patients in West Bengal was detected to possess resistance determinants against higher-generation cephalosporins depicting the picture of the community carriage (Saha et al., 2014), which indicates the milker’s hands as another probable source of infection. Indeed, a limitation of the study is the lack of sampling from the milkers, which could successfully establish the hypothesis on the origin and transmission of K. pneumoniae clones.
Among the ESBL-producing Klebsiella spp., 56 isolates (56/67, 83.6%) were also confirmed as AmpC β-lactamase producers. The AmpC β-lactamase producing bacteria are although less common in nature but co-existence of ESBL and AmpC β-lactamase is a significant concern, as the co-existence of both resistance types exhibits a broader resistance profile (EFSA, 2011). AmpC β-lactamase-producing Enterobacteriaceae were detected earlier in raw milk and milk products (cheese) in different countries (Endimiani et al., 2012; Özadam and Özpinar, 2016).
All the ESBL- and AmpC-producing Klebsiella spp. possessed blaCTX-M (67/67, 100%) and blaAmpC (56/56, 100%), respectively, in PCR. Additionally blaSHV (40/67, 59.7%) and blaTEM (38/67, 56.7%) were detected in Klebsiella isolates. None of the isolates were positive for the plasmid-mediated AmpC β-Lactamase gene (CITM). In total, 17 (17/67, 25.3%) ESBL-producing Klebsiella isolates were confirmed as Klebsiella pneumoniae in PCR. Similarly, the predominance of CTX-M was detected earlier in Enterobacteriaceae, including Klebsiella spp. isolated from raw milk, bulk tank milk and dairy cattle throughout the world (Badri et al., 2017; Dahmen et al., 2013; Odenthal et al., 2016; Schmid et al., 2013). In previous studies from the same geographical location, CTX-M was predominantly detected among the ESBL-producing E. coli in cattle, poultry and pigs (Bandyopadhyay et al., 2015; Kar et al., 2015; Samanta et al., 2015). Similarly, earlier studies in food animals and birds confirmed the distribution of blaAmpC in this location (Banerjee et al., 2019; Samanta et al., 2018).
The present study revealed a higher occurrence of class 1 integron in ESBL-producing Klebsiella spp. isolates (53.7%), depicting their high transmission potential.
All ESBL-producing Klebsiella sp. isolates were resistant to ceftazidime, cefepime and penicillin-G but sensitive to chloramphenicol. Higher resistance was observed against piperacillin (97%), amoxycillin/clavulanic acid (94%), ceftizoxime (86.5%) and cefoperazone (85%), whereas a lower degree of resistance was found against amikacin (1.5%), tobramycin (3%), gentamicin (3%), ceftriaxone/tazobactam (7.5%), ampicillin/sulbactam (9%), ciprofloxacin (13.4%) and levofloxacin (13.4%). Similarly, the Enterobacteriaceae isolated from milk and milk products showed phenotypical resistance against penicillin G, cloxacillin, ceftriaxone, cefotaxime, ceftazidime, tetracycline, ciprofloxacin and norfloxacin (Badri et al., 2017; Geser et al., 2012; Osman et al., 2014; Su et al., 2016). The MIC of cefotaxime, ceftazidime, and ceftriaxone against ESBL-producing Klebsiella sp. isolates varied from 0.01 µg/ml to 240 µg/ml, 3 µg/ml to >240 µg/ml and 0.01 µg/ml to 240 µg/ml, respectively (Table 2). All the isolates were found to be multidrug resistant based on the criterion of resistance to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012) (Table 2).
Table 2
Antimicrobial resistance profile, bla genotyping and MIC values of different antibiotics against ESBL-producing Klebsiella isolated from milk samples in West Bengal, India
| Isolate No. | Genotype | Antimicrobial Resistance Profile |
MICCTX (µg/ml) |
MICCAZ (µg/ml) |
MICCTR (µg/ml) |
| K1 | blaCTX-M blaTEM blaSHV blaAMPC | CPZ CX CPM P AMC CZX PI CAZ | 30 | >30 | >10 |
| K2 | blaCTX-M | CPM P PI CAZ | 30 | 30 | 5 |
| K3 | blaCTX-M | CPM P CZX CAZ | 10 | 30 | >30 |
| K4 | blaCTX-M blaTEM blaSHV | CPM P PI CAZ | 30 | 30 | 5 |
| K5 | blaCTX-M | CPM P AMC PI CAZ | 30 | >30 | >15 |
| K6 | blaCTX-M | ETP CPZ CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K7 | blaCTX-M | ETP CPZ CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K8 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT CPZ CX S CPM P AMC DO CZX PI CAZ | 30 | 120 | 30 |
| K9 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >30 | 30 |
| K10 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >30 | 30 |
| K11 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT CPZ CX S CPM P A/S AMC DO TOB CZX PI CAZ | 30 | >60 | >30 |
| K12 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >60 | >30 |
| K13 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >240 | >15 |
| K14 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM P AMC DO CZX PI CAZ | 30 | 120 | 30 |
| K15 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT CPZ CX S CPM P AMC DO CZX PI CAZ | 30 | 120 | 30 |
| K16 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT CPZ CX S CPM P AMC DO CZX PI CAZ | 30 | >30 | 30 |
| K17 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >30 | 30 |
| K18 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM P AMC DO CZX PI CAZ | 60 | 30 | 30 |
| K19 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT CPZ CX S CPM P AMC DO CZX PI CAZ | 60 | 30 | 30 |
| K20 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM P AMC CZX PI CAZ | 30 | >240 | >15 |
| K21 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT CPZ CX S CPM P AMC CZX PI CAZ | 30 | >240 | >15 |
| K22 | blaCTX-M blaSHV blaAMPC | CPZ CX CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K23 | blaCTX-M blaSHV blaAMPC | CPZ CX CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K24 | blaCTX-M blaAMPC | CPZ CX CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K25 | blaCTX-M blaAMPC | CPZ CX CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K26 | blaCTX-M blaTEM blaSHV blaAMPC | CPZ CX CPM P AMC CZX PI CAZ | 30 | 30 | 5 |
| K27 | blaCTX-M blaSHV blaAMPC | ETP CPZ CX CPM P AMC CZX PI CAZ | 10 | 30 | >30 |
| K28 | blaCTX-M blaTEM blaSHV blaAMPC intI | ETP CPZ CX CPM P AMC CZX PI CAZ | 10 | 30 | >30 |
| K29 | blaCTX-M blaSHV | ETP CPZ CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K30 | blaCTX-M blaSHV | ETP CPZ CPM P AMC CZX PI CAZ | 30 | 30 | 30 |
| K31 | blaCTX-M blaSHV | ETP CPZ CPM P AMC CZX PI CAZ | >30 | 30 | >240 |
| K32 | blaCTX-M blaTEM blaAMPC | CX CPM P AMC PI CAZ | 0.1 | 7.5 | 0.01 |
| K33 | blaCTX-M blaTEM blaAMPC intI sul1 | COT CPZ CX CPM P AMC CZX PI CAZ | 120 | 240 | 30 |
| K34 | blaCTX-M blaTEM blaAMPC intI sul1 | COT CPZ CX CPM P AMC DO CZX PI CAZ | 120 | 240 | 30 |
| K35 | blaCTX-M blaTEM blaSHV blaAMPC intI sul2 | COT ETP CIP CPZ CX CPM LE P AMC DO CZX PI CAZ | >30 | >240 | >240 |
| K36 | blaCTX-M blaSHV blaAMPC intI sul2 | COT ETP CIP CPZ CIT CX CPM TE LE P AMC DO CZX PI CAZ | >30 | >240 | >240 |
| K37 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 | COT CPZ CX CPM P AMC DO CZX PI CAZ | 120 | 240 | 30 |
| K38 | blaCTX-M blaSHV blaAMPC intI sul1 | COT ETP CPZ CIT CX CPM TE P AMC DO CZX PI CAZ | 120 | 240 | 30 |
| K39 | blaCTX-M blaSHV blaAMPC sul1 sul2 | COT CPZ CX CPM TE P A/S AMC DO CZX PI CAZ | 240 | 30 | 120 |
| K40 | blaCTX-M blaSHV blaAMPC sul1 sul2 | COT CPZ CX CPM TE P AMC DO TOB CZX PI CAZ | 240 | 30 | 120 |
| K41 | blaCTX-M blaSHV blaAMPC intI sul1 sul2 | COT ETP CIP CPZ CX CPM GEN TE LE P AMC DO CZX PI CAZ | 30 | >30 | >10 |
| K42 | blaCTX-M blaSHV blaAMPC intI sul1 sul2 | COT ETP CIP CPZ CX CPM GEN LE P AMC DO CZX PI CAZ | 30 | >30 | >10 |
| K43 | blaCTX-M blaTEM blaSHV blaAMPC intI qnrS sul1 | COT ETP CIP CPZ CX CPM LE P AMC DO CZX PI CAZ | 120 | 240 | 30 |
| K44 | blaCTX-M blaTEM blaSHV blaAMPC intI qnrS sul1 | COT ETP CIP CPZ CIT CX CPM LE P AMC DO CZX PI CAZ | 120 | 240 | 30 |
| K45 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX S CPM TE P A/S AMC CZX PI CAZ | 60 | 30 | 30 |
| K46 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM TE P A/S AMC DO CZX PI CAZ | 60 | 30 | 30 |
| K47 | blaCTX-M blaTEM blaAMPC intI qnrS sul1 sul2 | COT ETP CIP CPZ CX CPM TE LE P AMC DO CZX PI CAZ | >10 | 30 | 60 |
| K48 | blaCTX-M blaTEM blaAMPC intI qnrS sul1 sul2 | COT ETP CIP CPZ CX CPM TE LE P AMC DO CZX PI CAZ | >30 | 30 | >30 |
| K49 | blaCTX-M blaAMPC | ETP CPZ CX CPM TE P AMC DO CZX PI CAZ | 120 | 120 | 60 |
| K50 | blaCTX-M blaTEM blaAMPC intI qnrS sul1 sul2 | COT ETP CIP CPZ CX CPM TE P AMC DO CZX PI CAZ | >30 | 30 | >30 |
| KP1 | blaCTX-M blaSHV | ETP CPM P AMC DO PI CAZ | 10 | 30 | >30 |
| KP2 | blaCTX-M blaSHV blaAMPC | CX CPM AK P AMC CZX PI CAZ | 0.01 | 3 | 0.01 |
| KP3 | blaCTX-M blaSHV blaAMPC | ETP CX CPM P AMC PI CAZ | 30 | 30 | 5 |
| KP4 | blaCTX-M blaSHV | ETP CPM P AMC PI CAZ | 30 | >30 | 15 |
| KP5 | blaCTX-M blaAMPC | CX CPM P CAZ | 0.01 | 3 | 0.01 |
| KP6 | blaCTX-M blaSHV blaAMPC | CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >60 | >30 |
| KP7 | blaCTX-M blaSHV blaAMPC | CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >60 | >30 |
| KP8 | blaCTX-M blaSHV blaAMPC | CPZ CX CPM P AMC DO CZX PI CAZ | 30 | 30 | 30 |
| KP9 | blaCTX-M blaTEM blaSHV blaAMPC | CPZ CX CPM P AMC DO CZX PI CAZ | 30 | 30 | 30 |
| KP10 | blaCTX-M blaSHV blaAMPC | CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >30 | 30 |
| KP11 | blaCTX-M blaTEM blaSHV blaAMPC | CPZ CX CPM P AMC DO CZX PI CAZ | 30 | >30 | 30 |
| KP12 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT ETP CPZ CX S CPM P AMC CZX PI CAZ | 30 | 120 | 30 |
| KP13 | blaCTX-M blaTEM blaAMPC intI sul1 sul2 | COT ETP CPZ CX S CPM P AMC DO CZX PI CAZ | 30 | >240 | >15 |
| KP14 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM TE P AMC DO PI CAZ | 0.01 | 3 | 0.01 |
| KP15 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 | COT ETP CPZ CIT CX CPM TE LE P AMC CZX PI CAZ | 60 | 30 | 30 |
| KP16 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT ETP CPZ CX CPM TE P A/S AMC DO CZX PI CAZ | 60 | 60 | 30 |
| KP17 | blaCTX-M blaTEM blaSHV blaAMPC intI sul1 sul2 | COT ETP CPZ CIT CX S CPM TE P A/S AMC DO CZX PI CAZ | 60 | 60 | 30 |
However, none of the ESBL-producing Klebsiella spp. isolates were positive for carbapenemase and metallo-beta-lactamase production in phenotypic assays. Overuse or misuse of antibiotics is associated with the generation of resistance against the antibiotics used in a particular setting. The multi-drug resistance profile of ESBL-producing Klebsiella spp. isolates showed the pattern of antibiotic exposure (cephalosporins, enrofloxacin and tetracycline, not carbapenem) of the studied cattle. Moreover, possession of blaESBL genes in ESBL-producing Klebsiella sp. isolates indicated about the possession of single conjugative plasmid which may also carry the sul gene causing co-resistance against co-trimoxazole (Cantón and Coque, 2006).
Five out of the nine ciprofloxacin and/or levofloxacin resistant Klebsiella spp. isolates possessed plasmid-mediated quinolone resistance gene (qnrS). The MIC of ciprofloxacin against qnr positive Klebsiella spp. isolates was found to be >32 µg/ml (Table 2). Similarly, earlier studies showed an MIC of 33.3 µg/ml produced by qnrS1 possessing Enterobacteriaceae isolates (van der Putten et al., 2019). Among co-trimoxazole-resistant (n = 37) ESBL-producing Klebsiella sp. isolates, seven and two isolates were positive for the sul1and sul2 genes, respectively, whereas, 28 isolates possessed both the sul1and sul2, but none of the isolates were positive for sul3 in PCR.
All 67 ESBL-producing Klebsiella spp. isolates were typeable with the primers used in RAPD-PCR. The amplified fragment size ranged from 170 bp to 4178 bp (calculated by Doc-itLs image analysis software, UVP, UK). The phylogenetic analysis of the studied isolates revealed that the strains isolated from same district were grouped in same cluster, indicating their phylogenetic relationship (K6 and K7; K49 and K50; K8 and K14; K39 and K40; K37 and K44; KP7, KP8 and KP10; K9, K10, KP13 and K17; KP9 and KP11).
CONCLUSIONS
The present study revealed a moderately higher occurrence of multi-drug resistant ESBL-/AmpC-producing Klebsiella spp. in raw milk collected from healthy as well as infected cattle. The occurrence of ESBL-producing Klebsiella spp. was significantly higher in milk samples collected from cattle suffering with mastitis than in those from healthy ones. Among the ESBL-producing Klebsiella spp., 56 isolates were also detected to produce AmpC β-lactamases.
The present study detected the occurrence of class 1 integron in ESBL-producing Klebsiella spp. Isolates,87 depicting their high transmission possibility. The phylogenetic analysis of the studied isolates revealed that the strains isolated from same location had clonal relationship.
The study made the consumers aware to avoid the raw milk consumption to prevent the spread of antimicrobial resistance
Author Contributions: AM, JB, SG collected the samples and conducted the methodology part. SNJ and IS supervised the study. IS conceptualized the study and wrote the primary manuscript. SB and TKD edited the manuscript. All authors declare that they have read and approved the publication of the manuscript in the present form.
Acknowledgement: The authors provide sincere thanks to Honourable Vice Chancellor, West Bengal University of Animal and Fishery Sciences for the infrastructure and facilities.
Funding: There was no external funding for this study.
Conflicts of Interest: The authors declare that there is no conflict of interest.
REFERENCES
Ahmed, H.F.; Hanaa, A.E.; Azza, M.K.; Samah, F.D.; El-Magd, M.A.; Darwish, S.A.; El Saka, H.I. Phenotypic and molecular identification of Klebsiella and Salmonella species isolated from subclinical mastitis milk of Egyptian buffalo. Global Veterinaria. 2016, 16, 500-507. https://doi.org/10.5829/idosi.gv.2016.16.05.10390
Badri, A.M.; Ibrahim, I.T.; Mohamed, S.G.; Garbi, M.I.; Kabbashi, A.S.; Arbab, M.H. Prevalence of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae isolated from raw milk samples in Al Jazirah State, Sudan. Molecular Biology. 2017, 7, 201. https://doi.org/10.4172/2168-9547.1000201
Bandyopadhyay, S.; Samanta, I.; Bhattacharyya, D.; Nanda, P.K.; Kar, D.; Chowdhury, J.; Dandapat, P.; Das, A.K.; Batul, N.; Mondal, B.; Dutta, T.K. Co-infection of methicillin-resistant Staphylococcus epidermidis, methicillin-resistant Staphylococcus aureus and extended spectrum β-lactamase producing Escherichia coli in bovine mastitis-three cases reported from India. Veterinary Quarterly. 2015, 35, 56-61. https://doi.org/10.1080/01652176.2014.984365
Banerjee, A.; Bardhan, R.; Chowdhury, M.; Joardar, S.N.; Isore, D.P.; Batabyal, K.; Dey, S.; Sar, T.K.; Bandyopadhyay, S.; Dutta, T.K.; Samanta, I. Characterization of beta‐lactamase and biofilm producing Enterobacteriaceae isolated from organized and backyard farm ducks. Letters in Applied Microbiology. 2019, 69, 110-115. https://doi.org/10.1111/lam.13170
Birgy, A.; Bidet, P.; Genel, N.; Doit, C.; Decré, D.; Arlet, G.; Bingen, R. Phenotypic screening of carbapenemases and associated beta-lactamases in carbapenem-resistant Enterobacteriaceae. Journal of Clinical Microbiology. 2012, 50, 1295-1302. https://doi.org/10.1128%2FJCM.06131-11
Brisse, S.; Verhoef, J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. International Journal of Systematic and Evolutionary Microbiology. 2001, 51, 915-924. https://doi.org/10.1099/00207713-51-3-915
Cantón, R.; Coque, T.M. The CTX-M β-lactamase pandemic. Current Opinion in Microbiology. 2006, 9, 466-475. https://doi.org/10.1016/j.mib.2006.08.011
Cao, V.; Lambert, T.; Nhu, D.Q.; Loan, H.K.; Hoang, N.K.; Arlet, G.; Courvalin, P. Distribution of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae in Vietnam. Antimicrobial Agents and Chemotherapy. 2002, 46, 3739-3743. https://doi.org/10.1128/aac.46.12.3739-3743.2002
Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro surveillance. 2008, 13, 19044.
Dahmen, S.; Métayer, V.; Gay, E.; Madec, J.Y.; Haenni, M. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Veterinary Microbiology. 2013, 162, 793-799. https://doi.org/10.1016/j.vetmic.2012.10.015
Diab, M.; Hamze, M.; Bonnet, R.; Saras, E.; Madec, J.Y.; Haenni, M. OXA-48 and CTX-M-15 extended-spectrum beta-lactamases in raw milk in Lebanon: epidemic spread of dominant Klebsiella pneumoniae clones. Journal of Medical Microbiology. 2017, 66, 1688-1691. https://doi.org/10.1099/jmm.0.000620
EFSA. Panel on Biological Hazards: Scientific Opinion on the public health risks of bacterial strains producing extended‐spectrum β‐lactamases and/or AmpC β‐lactamases in food and food‐producing animals. EFSA Journal. 2011, 9, 2322. https://doi.org/10.2903/j.efsa.2011.2322
El-Sukhon, S.N. Identification and characterization of Klebsiellae isolated from milk and milk products in Jordan. Food Microbiology. 2003, 20, 225-230. https://doi.org/10.1016/S0740-0020(02)00085-0
Endimiani, A.; Rossano, A.; Kunz, D.; Overesch, G.; Perreten, V. First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland. Diagnostic Microbiology and Infectious Disease. 2012, 73, 31-38. https://doi.org/10.1016/j.diagmicrobio.2012.01.004
Féria, C.; Ferreira, E.; Correia, J.D.; Gonçalves, J.; Caniça, M. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. Journal of Antimicrobial Chemotherapy. 2002, 49, 77-85. https://doi.org/10.1093/jac/49.1.77
Frank, T.; Gautier, V.; Talarmin, A.; Bercion, R.; Arlet, G. Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). Journal of Antimicrobial Chemotherapy. 2007, 59, 742-745. https://doi.org/10.1093/jac/dkl538
Geser, N.; Stephan, R.; Hächler, H. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Veterinary Research. 2012, 8, 21. https://doi.org/10.1186/1746-6148-8-21
Gundogan, N.; Yakar, U.A. Siderophore production, serum resistance, hemolytic activity and extended‐spectrum β‐lactamase‐producing Klebsiella species isolated from milk and milk products. Journal of Food Safety. 2007, 27, 251-264. https://doi.org/10.1111/j.1745-4565.2007.00077.x
Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; Brisse, S. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. PNAS. 2015, 112. https://doi.org/10.1073/pnas.1501049112
Jayaweera, T.S.P.J.; Randika, J.L.P.C.; Gamage, H.G.C.L.; Udawatta, N.N.; Ellepola, W.U.N.T.S.; Ruwanjith, H.A.D.; Ruwandeepika, H.A.D. Isolation of Escherichia coli, Klebsiella spp. and Staphylococcus spp. from bovine mastitic milk in NuweraEliya District of Sri Lanka and their sensitivity to commonly used antibiotics. South Asian Journal of Research in Microbiology. 2018, 1-13. https://doi.org/10.9734/sajrm/2018/v1i2766
Kar, D.; Bandyopadhyay, S.; Bhattacharyya, D.; Samanta, I.; Mahanti, A.; Nanda, P.K.; Mondal, B.; Dandapat, P.; Das, A.K.; Dutta, T.K.; Bandyopadhyay, S. Molecular and phylogenetic characterization of multidrug resistant extended spectrum beta-lactamase producing Escherichia coli isolated from poultry and cattle in Odisha, India. Infection, Genetics and Evolution. 2015, 29, 82-90. https://doi.org/10.1016/j.meegid.2014.11.003
Landes, M.; Cessna, J.; Kuberka, L.; Jones, K. India’s Dairy Sector: Structure, Performance, and Prospects, A report from the economic research service. United States Department of Agriculture, LDPM-272-01, 2017.
Liebana, E.; Carattoli, A.; Coque, T.M.; Hasman, H.; Magiorakos, A.P.; Mevius, D.; Peixe, L.; Poirel, L.; Schuepbach-Regula, G.; Torneke, K.; Torren-Edo, J. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clinical Infectious Diseases. 2013, 56, 1030-1037. https://doi.org/10.1093/cid/cis1043
Lim, K.T.; Yeo, C.C.; Yasin, R.M.; Balan, G.; Thong, K.L. Characterization of multidrug-resistant and extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains from Malaysian hospitals. Journal of Medical Microbiology. 2009, 58, 1463-1469. https://doi.org/10.1099/jmm.0.011114-0
Liu, Y.; Liu, C.; Zheng, W.; Zhang, X.; Yu, J.; Gao, Q.; Hou, Y.; Huang, X. PCR detection of Klebsiella pneumoniae in infant formula based on 16S–23S internal transcribed spacer. International Journal of Food Microbiology. 2008, 125, 230-235. https://doi.org/10.1016/j.ijfoodmicro.2008.03.005
Locatelli, C.; Scaccabarozzi, L.; Pisoni, G.; Moroni, P. CTX-M1 ESBL-producing Klebsiella pneumoniae subsp. pneumoniae isolated from cases of bovine mastitis. Journal of Clinical Microbiology. 2010, 48, 3822-3823. https://doi.org/10.1128/jcm.00941-10
Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; Paterson, D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2012, 18, 268-281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrobial Agents and Chemotherapy. 2000, 44, 1568-1574. https://doi.org/10.1128/aac.44.6.1568-1574.2000
NMC (National Mastitis Council). Microbiological procedures for the diagnosis of udder infection(3rded.), Arlington, VA: National Mastitis Council Inc, 1990.
Odenthal, S.; Akineden, Ö.; Usleber, E. Extended-spectrum β-lactamase producing Enterobacteriaceae in bulk tank milk from German dairy farms. International Journal of Food Microbiology. 2016, 238, 72-78. https://doi.org/10.1016/j.ijfoodmicro.2016.08.036
Ohnishi, M.; Okatani, A.T.; Harada, K.; Sawada, T.; Marumo, K., Murakami, M.; Sato, R.; Esaki, H.; Shimura, K., Kato, H.; Uchida, N. Genetic characteristics of CTX-M-type extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae involved in mastitis cases on Japanese dairy farms, 2007 to 2011. Journal of Clinical Microbiology. 2013, 51, 3117-3122. https://doi.org/10.1128/jcm.00920-13
Osman, K.M.; Hassan, H.M.; Orabi, A.; Abdelhafez, A.S. Phenotypic, antimicrobial susceptibility profile and virulence factors of Klebsiella pneumoniae isolated from buffalo and cow mastitic milk. Pathogens and Global Health. 2014, 108, 191-199. https://doi.org/10.1179%2F2047773214Y.0000000141
Özadam, A.; Özpinar, H. Phenotypic determination of ESBL-and AmpC-producing Enterobacteriaceae in cheese samples. International Journal of Food Engineering Research. 2016, 2, 59-70.
Özpınar, H.; Tekiner, İ.H.; Sarıcı, B.; Çakmak, B.; Gökalp, F.; Özadam, A. Phenotypic characterization of ESBL-and AmpC-Type betalactamases in Enterobacteriaceaefrom chicken meat and dairy products. Ankara Üniversitesi Veteriner Fakültesi Dergisi. 2017, 64, 267-272. https://doi.org/10.1501/Vetfak_0000002809
Quinn, P.J.; Carter, M.E.; Markey, B.K.; Carter, G.R. Clinical Veterinary Microbiology. London, Wolfe Publishing, 1994.
Patel, J.B.; Cockerill, F.R.; Alder, J.; Bradford, P.A.; Eliopoulos, G.M.; et al. Performance standards for antimicrobial susceptibility testing: Twenty-fourth informational supplement, M100-S24, 34, Clinical and Laboratory Standards Institute (CLSI) West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA, 2014.
Parekh, T.S.; Subhash, R. Molecular and bacteriological examination of milk from different milch animals with special reference to coliforms. Current Research in Bacteriology. 2008, 1, 56-63. http://dx.doi.org/10.3923/crb.2008.56.63
Saha, S.; Nayak, S.; Bhattacharyya, I.; Saha, S.; Mandal, A.; Chakraborty, S.; Bhattacharyya, R.; Chakraborty, R.; Franco, O.L.; Mandal, S.M.; Basak, A. Understanding the patterns of antibiotic susceptibility of bacteria causing urinary tract infection in West Bengal, India. Frontiers in Microbiology. 2014, 5, 463. https://doi.org/10.3389/fmicb.2014.00463
Samanta, I.; Joardar, S.N.; Mahanti, A.; Bandyopadhyay, S.; Sar, T.K.; Dutta, T.K. Approaches to characterize extended spectrum beta-lactamase/beta-lactamase producing Escherichia coli in healthy organized vis-a-vis backyard farmed pigs in India. Infection, Genetics and Evolution. 2015, 36, 224-230. https://doi.org/10.1016/j.meegid.2015.09.021
Samanta, A.; Mahanti, A.; Chatterjee, S.; Joardar, S.N.; Bandyopadhyay, S.; Sar, T.K.; Mandal, G.P.; Dutta, T.K.; Samanta, I. Pig farm environment as a source of beta-lactamase or AmpC-producing Klebsiella pneumoniae and Escherichia coli. Annals of Microbiology. 2018, 68, 781-791. https://doi.org/10.1007/s13213-018-1387-2
Schmid, A.; Hörmansdorfer, S.; Messelhäusser, U.; Käsbohrer, A.; Sauter-Louis, C.; Mansfeld, R. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli on Bavarian dairy and beef cattle farms. Applied and Environmental Microbiology. 2013, 79, 3027-3032. https://doi.org/10.1128%2FAEM.00204-13
Su, Y.; Yu, C.Y.; Tsai, Y.; Wang, S.H.; Lee, C.; Chu, C.: Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli from the milk of cows with clinical mastitis in Southern Taiwan. Journal of Microbiology, Immunology and Infection. 2016, 49, 892-901. https://doi.org/10.1016/j.jmii.2014.10.003
Skočková, A.; Bogdanovičová, K.; Koláčková, I.; Karpíšková, R.: Antimicrobial-resistant and extended-spectrum β-lactamase–producing Escherichia coli in raw cow’s milk. Journal of Food Protection. 2015, 78, 72-77. https://doi.org/10.4315/0362-028x.jfp-14-250
Tan, T.Y.; Ng, L.S.Y.; He, J.; Koh, T.H.; Hsu, L.Y. Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrobial Agents and Chemotherapy. 2009, 53, 146-149. https://doi.org/10.1128/aac.00862-08
Tepeli, S.Ö.; Zorba, N.N.D. Frequency of extended-spectrum β-lactamase (ESBL)–and AmpC β-lactamase–producing Enterobacteriaceae in a cheese production process. Journal of Dairy Science. 2018, 101, 2906-2914. https://doi.org/10.3168/jds.2017-13878
Van, T.T.H.; Chin, J.; Chapman, T.; Tran, L.T.; Coloe, P.J.: Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. International Journal of Food Microbiology. 2008, 124, 217-223. https://doi.org/10.1016/j.ijfoodmicro.2008.03.029
van der Putten, B.C.; Remondini, D.; Pasquini, G.; Janes, V.A.; Matamoros, S.; Schultsz, C. Quantifying the contribution of four resistance mechanisms to ciprofloxacin MIC in Escherichia coli: a systematic review. Journal of Antimicrobial Chemotherapy. 2019, 74, 298-310. https://doi.org/10.1093/jac/dky417
Weill, F.X.; Lailler, R.; Praud, K.; Kérouanton, A.; Fabre, L.; Brisabois, A.; Grimont, P.A.; Cloeckaert, A. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. Journal of Clinical Microbiology. 2004, 42, 5767-5773. https://doi.org/10.1128%2FJCM.42.12.5767-5773.2004
Academic Editor: Prof. dr. Daniel Simeanu
Publisher Note: Regarding jurisdictional assertions in published maps and institutional affiliations ALSE maintain neutrality.
Bandyopadhyay Samiran, Banerjee Jaydeep, Dutta Tapan Kumar, Ghosh Sarbaswarup, Joardar Siddhartha Narayan, Mahanti Achintya, Samanta Indranil



