Florin-Daniel Lipșa, Rod Snowdon, Benjamin Wittkop, Wolfgang Friedt
ABSTRACT. Rapeseed meal, a by-product of oilseed extraction related to the agri-food and biofuel industries due to its favourable composition of essential amino acids, is currently utilised for animal feed. In this study, 166 doubled haploid (DH) rapeseed lines from the segregating Brassica napus doubled haploid population YE2-DH were genetically and chemically analysed for phenolic acids. The relationship between seed colour and phenolic acid fractions in B. napus was investigated using these analyses to improve the quality of rapeseed meal. High-performance liquid chromatography assays were used to estimate phenolic acid content, and the outcomes were used to identify quantitative trait loci (QTL). Nine quantitative feature loci for three distinct phenolic acid compounds were mapped to seven linkage groups. A minor QTL for sinapine was located on linkage group C05 in the same interval as the QTL for seed colour. On chromosome A09, two loci for phenolic acids colocalised with the main QTL for seed colour. Closely linked molecular markers for the target traits (seed colour, phenolic acids) identified during this study could be useful tools for breeding new oilseed rape cultivars with lower levels of antinutritive compounds.
Keywords: Brassica napus; phenolic acids; meal quality; QTL mapping.
Cite
ALSE and ACS Style
Lipșa, F.-D.; Snowdon, R.; Wittkop, B.; Friedt, W. Quantitative genetic analysis of phenolic acids in oilseed rape meal. Journal of Applied Life Sciences and Environment 2022, 55 (2), 133-144.
https://doi.org/10.46909/alse-552051
AMA Style
Lipșa FD, Snowdon R, Wittkop B, Friedt W. Quantitative genetic analysis of phenolic acids in oilseed rape meal. Journal of Applied Life Sciences and Environment. 2022; 55 (2): 133-144.
https://doi.org/10.46909/alse-552051
Chicago/Turabian Style
Lipșa, Florin-Daniel, Rod Snowdon, Benjamin Wittkop, and Wolfgang Friedt. 2022. “Quantitative genetic analysis of phenolic acids in oilseed rape meal” Journal of Applied Life Sciences and Environment 55, no. 2: 133-144.
https://doi.org/10.46909/alse-552051
View full article (HTML)
Quantitative Genetic Analysis of Phenolic Acids in Oilseed Rape Meal
Florin-Daniel LIPȘA1*, Rod SNOWDON2, Benjamin WITTKOP2 and Wolfgang FRIEDT2
1 Iasi University of Life Sciences, Faculty of Agriculture, Department of Food Technologies, 3, Mihail Sadoveanu Alley, 700490, Iasi, Romania;
2 Justus Liebig University, Department of Plant Breeding, Heinrich-Buff-Ring 26-32, 35392-Giessen, Germany; e-mail: rod.snowdon@agrar.uni-giessen.de, benjamin.wittkop@agrar.uni-giessen.de, wolfgang.friedt@agrar.uni-giessen.de
*Correspondence: flipsa@uaiasi.ro
Received: Nov. 24, 2022. Revised: Dec. 13, 2022. Accepted: Dec. 13, 2022. Published online: Jan. 16, 2023
ABSTRACT. Rapeseed meal, a by-product of oilseed extraction related to the agri-food and biofuel industries due to its favourable composition of essential amino acids, is currently utilised for animal feed. In this study, 166 doubled haploid (DH) rapeseed lines from the segregating Brassica napus doubled haploid population YE2-DH were genetically and chemically analysed for phenolic acids. The relationship between seed colour and phenolic acid fractions in B. napus was investigated using these analyses to improve the quality of rapeseed meal. High-performance liquid chromatography assays were used to estimate phenolic acid content, and the outcomes were used to identify quantitative trait loci (QTL). Nine quantitative feature loci for three distinct phenolic acid compounds were mapped to seven linkage groups. A minor QTL for sinapine was located on linkage group C05 in the same interval as the QTL for seed colour. On chromosome A09, two loci for phenolic acids colocalised with the main QTL for seed colour. Closely linked molecular markers for the target traits (seed colour, phenolic acids) identified during this study could be useful tools for breeding new oilseed rape cultivars with lower levels of antinutritive compounds.
Keywords: Brassica napus; phenolic acids; meal quality; QTL mapping.
INTRODUCTION
Due to its favourable composition of essential amino acids, oilseed rape/canola (Brassica napus L., 2n=38, genome AACC) is a good source of vegetable protein (Yang et al., 2015; Cheng et al., 2022). In this context, fibre, dark-coloured tannins and bitter-tasting sinapate esters are of particular importance (TUM, 2019).
Phenolic acids and condensed tannins represent the predominant phenolic compounds in rapeseed, the total content of which is much higher than that in other oilseeds (soybean, cottonseed, peanut). They are responsible for the dark colour, bitter taste, unpleasant odour and astringent properties of rapeseed meal products due to the reduction in protein digestibility caused by their complex formation (Lipsa et al., 2012, Yang et al., 2022).
Phenolic acids are present in rapeseed in free, esterified and insoluble-bound forms and represent benzoic and cinnamic acid derivatives. Soluble phenolic compounds accumulate in rapeseed, mainly in the cotyledons. Sinapic acid accounts for the majority of free phenolic acids, up to 15% of total phenolics in rapeseed meal, and up to 99% of phenolic acids produced from esters and glucosides (Krygier et al., 1982; Naczk et al., 1998; Reungoat et al., 2022).
In terms of quantity, the esterified phenolic acids represent the largest proportion, with the choline ester of sinapic acid, sinapine, accounting for between 70% (Kolodziejczyk et al., 1999) and 80% (Obied et al., 2013; Aludatt et al., 2017) of all sinapic acid esters occurring in rapeseed and can reach contents in rapeseed meal of 1–2% (Rezaeizad et al., 2011).
A reduction in the phenolic compound content is therefore an indispensable prerequisite for the use of rapeseed meal in animal nutrition and in the human protein supply. Extensive studies on the sinapic acid ester content of various Brassica species and cultivars have shown considerable variability in this trait (Kozlowska et al., 1990; Milkowski and Strack, 2010). Rapeseed meal has a sinapine content that varies from 5 to 30 mg/g depending on the cultivar, growing environment and location (Shahidi and Naczk, 1995; Tan et al., 2011).
Although the available cultivars sometimes show significant differences in phenolic content and sinapine concentration, the development of newer cultivars with reduced sinapine content by classical breeding has been less successful and very time consuming. A promising way to reduce sinapine content is the genetic suppression of the two key enzymes, UDP-glucose:sinapic acid glucosyltransferase (SGT) and sinapoylglucose:choline sinapoyltransfe rase (sinapine synthase; SCT), in the sinapine biosynthesis pathway using antisense regulation (Milkowski et al., 2004; Corso et al., 2020). Such targeted genetic engineering approaches provide hope that the levels of phenolic compounds in canola can be sustainably reduced. An improvement in the nutritional value and possibly an increased use of rapeseed meal products in animal and human nutrition would be the result. Other ways to reduce phenolic acids from rapeseed meal are the use of biological methods, such as enzyme addition and microbial fermentation, to remove antinutritional factors. Enzyme-assisted methods and microbial fermentation have shown outstanding effects on the removal of antinutritional factors and are also considered potential strategies for rapeseed commercial feed production (Yang et al., 2022).
To identify closely linked molecular markers associated with quantitative trait loci involved in the acquisition and content variability of phenolic acids in rapeseed meal, the purpose of this study was to establish screening and analysis techniques for these compounds in B. napus seed from a doubled haploid population with 166 lines.
MATERIALS AND METHODS
The materials and methods are described in detail in a previous article of the authors (Lipsa et al., 2012). For a better understanding of the steps that are followed in the present study we present the structure of the chapter and the most important data for the genetic analysis of phenolic acids.
Plant material
A B. napus population of 166 doubled haploid lines, descended from a cross between the true-breeding line ‘1012/98’ (yellow seeds) and the German winter oilseed rape cultivar ‘Express’ (black seeds), was used to create a genetic map. Both parental lines presented low erucic acid and glucosinolate content in the seed (00 quality).
To conduct a quantitative trait loci (QTL) study of seed colour and related features, the population was cultivated over three seasons in various sites. The plants deposition and seed storage are similar with the those presented by Lipsa et al. (2012).
Seed colour and phenolic acid content evaluation
Utilising the NIR System 6500 and WinISI II software, seed colour was quantified based on levels of digital reflectance utilising near-infrared reflectance spectroscopy (NIRS) as describe by Lipsa et al. (2012).
Sample preparation and extraction of phenolic acid fractions from seed meal
The sample preparation for phenolic acid is similar with sample preparation for condensed tannins (Lipsa et al., 2012) Then the dried rapeseed meal samples (500 mg, 3 repetitions per line) were extracted twice for better results. sing solid-phase extraction (SPE) techniques modified from Naczk and Shahidi (2004) and Sun et al. (2006), the phenolic acids were eluted with water after crude rapeseed meal extract was added to activated, preconditioned C18 mini-columns
Chromatographic conditions and HPLC quantification of proanthocyanidins
For the quantification of the phenolic acids via HPLC, procyanidin B2 (Extrasynthese, France) was used as the internal standard. Using a Merck-Hitachi low-pressure gradient system, HPLC analysis for the quantification of phenolic acid fractions from all 166 DH lines was carried out
Genetic linkage map and QTL analysis
Genetic linkage map was created using a crucial logarithm of odds (LOD) score of 3.0 and a maximum likelihood distance of 35 cM. TL analysis was carried out using the composite interval mapping technique.
RESULTS
Phenolic acid content and seed colour variation
In the phenolic acid fraction (Figure 1), two major peaks were detected at retention times of 12.72 (F1PA2) and 14.37 min (F1PA1). These peaks have been identified by the co-migration of standard substances and probably correspond to sinapoylglucose and sinapine, the major phenolic compounds in rapeseed (Mabon et al., 1999; Vuorela et al., 2003; Milkowski et al., 2004; Zum Felde, 2007). Furthermore, two signals were detected at retention times of 19.47 (F1PA3) and 20.19 min (F1PA4). Phenotype information for various phenolic acid fractions and seed colour for all 166 DH lines, along with a summary of their unique features, are presented in Table 1. The genotypes of the double haploid population’s seed colours ranged from 2.9 (bright yellow) to 8.1 (black) in terms of optical light reflectance, while the phenolic acid content ranged from 601.7 to 1466.1 mg/100 g of oil-free rapeseed meal.
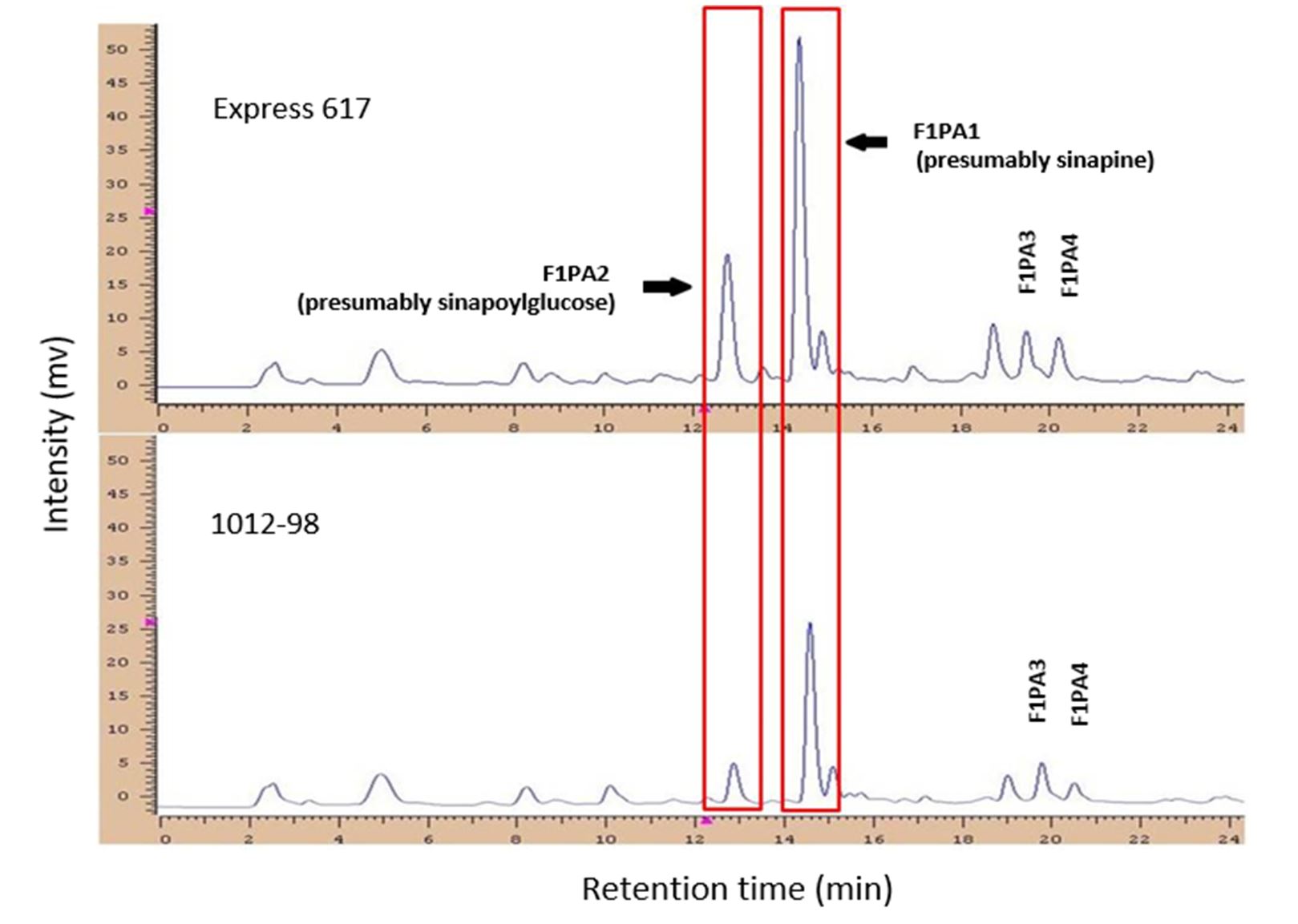
Figure 1 – High-performance liquid chromatography chromatograms (280 nm) of phenolic acid fractions from the black- and yellow-seeded parents (‘Express 617’ and ‘1012-98,’ respectively). The red areas show the major peaks for the phenolic acids, presumably sinapoylglucose and sinapine
Table 1
Seed colour and total and individual phenolic acid contents for the YE2 double haploid population and mean values from the yellow- (‘1012-98’) and black-seeded parental lines (‘Express 617’)
|
Seed trait or compound |
Min. |
Max. |
Mean |
SD |
Parental lines |
||
|
1012-98 |
Express 617 |
||||||
|
Seed colour1) |
2.9 |
8.1 |
6.0 |
1.1 |
2.8 |
8.3 |
|
|
Total phenolic acids2) |
601.7 |
1466.1 |
1012.5 |
164.8 |
352.5 |
968.5 |
|
|
Phenolic acids2) |
F1PA1 (pr. Sinapine) |
376.2 |
936.1 |
629.2 |
106.5 |
239.0 |
636.5 |
|
F1PA2 (pr. Sinapoylglucose) |
39.0 |
359.6 |
178.8 |
58.9 |
70.9 |
202.5 |
|
|
F1PA3 |
50.0 |
247.3 |
123.0 |
44.0 |
60.8 |
80.4 |
|
|
F1PA4 |
9.0 |
179.7 |
81.4 |
36.2 |
43.9 |
69.0 |
|
1) Seed colour as absorbance value (1 = white, 10 = black)
2) Phenolic acid content in mg/100 g dried rapeseed meal
pr – presumably
The most predominant phenolic compound was represented by sinapine (F1PA1), with a quantity ranging from 376.2 to 936.1 mg/100g dried meal, followed by sinapoylglucose (F1PA2) with 39.0 to 359.6 mg/100g defatted meal. The minimum content among the phenolic acids was found in the case of an unidentified peak F1PA4 with 9.0 to 179.7 mg/100 g of oil-free meal.
No significant correlation was found between seed colour and phenolic acid fractions F1PA2 (presumably sinapoylglucose), F1PA3 and F1PA4. A weak correlation (r=0.37*) was found between seed colour and phenolic acid F1PA1 (presumably sinapine). There are no data in the literature on unexpected correlations between the different phenolic acids and the mentioned traits. The largest proportion of phenolic acids in rapeseed was represented by sinapic acid or its esters (70–85%). These were predominantly found in the cotyledons and only as traces in the seed coat (0.06–0.24%). Among the phenolic acids, F1PA1 was correlated with F1PA2, and F1PA2 was correlated with both F1PA3 and F1PA4.
QTL analysis
Each QTLs’ contribution to phenotypic variance is shown in Table 2, along with its chromosomal location, LOD score and additive effect (partial R2).
Localisation of QTL for seed colour
For the seed colour trait, three QTLs were localised on linkage groups A09, C01 and C05 (Figure 2, Table 2), explaining a total of 66.8% of the phenotypic variance (R2). The positive alleles of all three QTLs originated from the yellow-seeded parent ‘1012-98.’ There was a highly significant QTL on linkage group N9, with a maximum LOD value of 6.4 and an additive effect of 0.45%. This QTL explained 40.9% of the observed partial phenotypic variance (part. R²) and played an important role in phenotype expression.
The QTL in linkage group N11 (LOD=7.6) had a smaller effect on phenotype expression (R2=19.0%; part. R2=19.8%) with an additive effect of 0.29%, while the QTL on linkage group N15 (LOD=3.9) explained only 12.2% of the observed partial phenotypic variance, with an additive effect of 0.23%.
Localisation of QTL for phenolic acids
For three different phenolic acid compounds (F1PA1, F1PA2, F1PA3), nine quantitative feature loci were mapped to seven linkage groups (Figure 2).
For trait F1PA1 (presumably sinapine), two major QTL (R² ≥ 10%) were located on linkage groups C01 and C05. These two QTLs had a clearly comparable effect on phenotype expression, with 13.6 and 12.8%. The two QTLs explained 21.1% of the phenotypic variance, and the LOD curves reached two maxima of 5.3 and 4.9, respectively.
For trait F1PA2 (presumably sinapoylglucose), three QTLs were localised to linkage groups A02, A09 and C01 at a LOD threshold of 3.0. The total phenotypic variance corresponded to 39.7%. The individual QTL explained 11.2 (A09), 16.8 (C01) and 40.0% (A02) of the observed phenotypic variance (R²), and 5.6 (A09), 14.9 (C01) and 32.2% (A02) of the partial phenotypic variance (part. R²). LOD values differed between 4.2 for linkage group A09 and 18.4 for linkage group A02.
The individual QTL for trait F1PA3 described between 8.8 and 16.0% of the observed phenotypic variance in the mapping population. Overall, 34.2% of the phenotypic variance was explained by these four QTLs. The locus with the highest partial phenotypic variance (part. R²=6.4%) and a LOD value of 6.3 was located on linkage group C03. Linkage group A09 contained the second QTL (LOD=4.9; part. R2=12.1%); the third was on linkage group C09 (LOD=4.4; part. R2=8.6%), and the fourth was on linkage group C06 (LOD=3.3; part. R2=8.6%).
Chromosomal QTL distribution in the YE2-DH population
In linkage group A02 (Figure 2), the main QTL for F1PA2 content (presumably sinapoylglucose) explained 40% of the phenotypic variance for these traits and had a statistically significant LOD value. The QTL on A02 had a very large effect on antinutritive phenolic acids but did not have any effect on seed colour because phenolic acids were predominantly localised in the cotyledons. For trait F1PA2, a total of three QTLs were localised on linkage groups A02, A09 and C01 at a LOD threshold of 3.0.
In linkage group A09, a 10 cM region with a strong effect on two different seed traits was detected. Here, the QTL for seed colour and phenolic acids (F1PA2, F1PA3) were colocalised. Among all QTL, the one with the strongest effect on seed colour explained 40.9% of the observed partial phenotypic variance (part. R²) and played an important role in phenotype expression.
In the same place as the QTL for seed colour on linkage group A09, there was a major QTL for ADF (acid detergent fibre), NDF (neutral detergent fibre) and ADL (acid detergent lignin) content (Bekele, 2008).
Table 2
A summary of the quantitative trait loci in the double haploid population’s genetic map for seed colour and phenolic acid components
|
Trait |
Marker interval |
Chromosome position (cM) |
LOD Score |
R2 (%) |
Partial R2 (%) |
|
|
Seed colour |
Ni4D09 / SSR1_144 |
A09 / 26 |
6.38*** |
16.2 |
40.9 |
|
|
E32M62_197 / E35M62_151 |
C01 / 48 |
7.61*** |
19.0 |
19.8 |
||
|
Ol10D02_A / E35M60_153 |
C05 / 4 |
3.89* |
10.2 |
12.2 |
||
|
Phenolic acids |
F1PA1 (presumably sinapine) |
tt1-1 / mr000228 |
C01 / 36 |
5,25** |
13,6 |
12,5 |
|
Ol10D02_A / E35M60_153 |
C05 / 6 |
4.94** |
12.8 |
11.6 |
||
|
F1PA2 (presumably sinapoylglucose) |
E35M60_305 / sn012964 |
A02 / 36 |
18,39*** |
40,0 |
32,2 |
|
|
Ni4D09 / SSR1_144 |
A09 / 24 |
4,24** |
11,2 |
5,6 |
||
|
tt1-1 / mr000228 |
C01 / 36 |
6,65*** |
16,8 |
14,9 |
||
|
F1PA3 |
Ni4D09 / SSR1_144 |
A09 / 24 |
4,91** |
12,7 |
12,1 |
|
|
Na10E02 / E40M60_334 |
C03 / 16 |
6,25*** |
15,9 |
16,4 |
||
|
E33M59_271 / Ol12E03 |
C06 / 56 |
3,32 |
8,8 |
8,6 |
||
|
E35M62_169 / E33M48_186 |
C09 / 38 |
4,38** |
11,4 |
8,6 |
||
* significant at p=0.05 by permutation analysis; ** significant at p=0.01; *** significant at p =0.001 R2 (%): phenotypic variation; Partial R2 (%): partial phenotypic variation
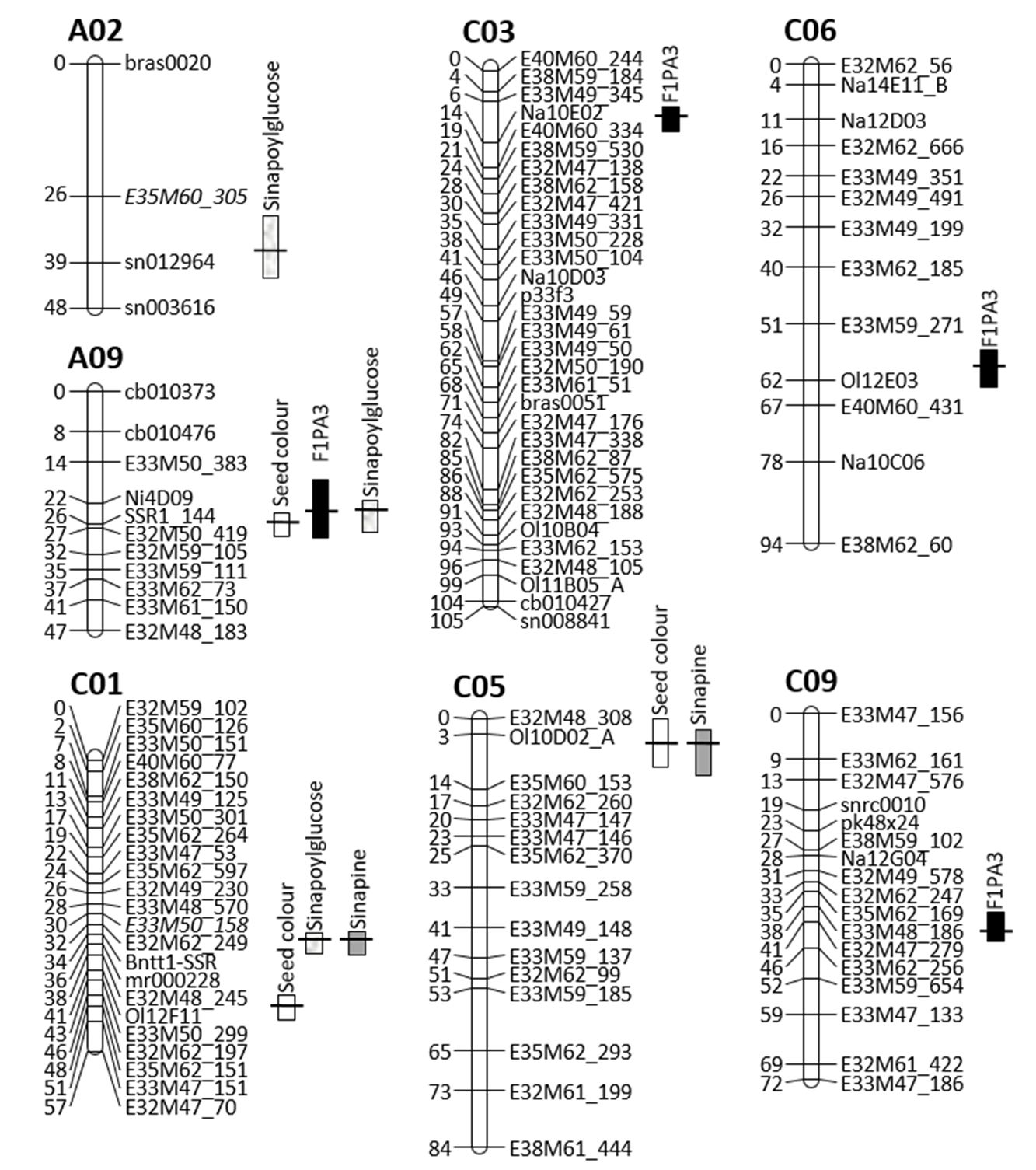
Figure 2 – Localisation of QTL for seed colour and phenolic acids on Brassica napus chromosomes of the mapping population
The SSR marker tt1_1 was located in the middle of linkage group NC01 of the population Express 617 × 1012-98 and was potentially tightly coupled to the tt1 gene in B. napus (Stein et al., 2013). The QTL for F1PA1 (putative sinapin) and F1PA2 (putative sinapoylglucose) content colocalised and were located on linkage group C01 in close proximity to marker tt1_1. The major QTL for the phenolic acid F1PA3 was located in linkage group C03 and explained 15.9% of the phenotypic variance. Figure 2 shows linkage group C05 of the B. napus DH population. In this linkage group, QTLs were distributed practically along the entire chromosome, influencing the quality traits of the canola seeds. The mapped QTL for seed colour and phenolic acid F1PA1 (presumably sinapine) colocalised in the upper part of the chromosome. In the middle region of chromosome C05, the main QTL for oil content was localised (data not shown in the paper). In linkage groups C06 and C09, two QTLs for F1PA3 content were detected, explaining 8.8 and 11.4% of the phenotypic variance.
DISCUSSION
The use of rapeseed meal or rapeseed extraction meal as a high-quality protein source for animal and human nutrition is an alternative to soybean meal. However, the use of rapeseed extraction meal as a complete feed is still limited due to antinutritive or undesirable ingredients (glucosinolates, sinapins, tannins and fibre components), which impair digestibility and protein availability and are also responsible for a bitter taste, especially when used in the feeding of monogastric livestock (chicken, pig) (Cheng et al., 2022).
Breeding and selection of yellow-seeded canola genotypes represents the opportunity to reduce antinutritional phenolic substances, while increasing oil and/or protein content, and to achieve better digestibility and protein availability, as well as higher acceptance of canola meal as animal feed.
In the course of the present work, 166 lines of a DH population from the cross Express 617 (black-seeded) × 1012-98 (yellow-seeded) were analysed for total and individual phenolic acid content using an HPLC assay and, as expected, a wide variation in quantification was observed.
Phenolic acids represent the predominant phenolic compounds in canola seed, with sinapine accounting for approximately 85% of all occurring sinapic acid esters in canola (Qiao and Classen, 2003). HPLC examination of a crude methanolic extract revealed two major peaks. With reference to Qiao and Classen (2003), these peaks are thought to correspond to sinapine (peak F1PA1) and sinapoylglucose (peak F1PA2), the two most abundant phenolic acids in canola meal. QTL analysis showed that the major QTL for F1PA1 content was located at the same position on chromosome C01, with a locus for F1PA2 content. In other words, there is probably a gene mutation at this location in the genome that results in a simultaneous reduction of both phenolic acids in the yellow-seeded parent. At the same locus, there are major QTL for total flavonoid and proanthocyanidin content and adjacent QTL for seed colour and oligomeric (F2PA3) and polymeric proanthocyanidins (F3PA4) (Lipsa et al., 2012). Because a copy of the potential candidate gene tt1 is also suspected here because of a neighbouring SSR marker (Bntt1), a mutation in a tt1 gene copy would be a possible explanation for these QTL. tt1 is a WIP zinc finger transcription factor with direct effects on endothelial cell formation. Mutations in this gene in Arabidopsis are expressed by a thin, transparent seed coat.
A minor QTL for F1PA1 content (presumably sinapin) is located on linkage group C05 in the same interval (2–8 cM) with the QTL for seed colour. On chromosome A09, two loci for F1PA2 and F1PA3 colocalised with the main QTL for seed colour (part. R2 = 40.9%), together with additional QTL for ADF, NDF and ADL content, and with a QTL with a small effect on flavonoid content (Bekele, 2008; Lipsa et al., 2012; Gacek et al., 2021). With regard to breeding for improvement of rapeseed meal quality, this result is significant because it demonstrates an association between seed coat thickness, phenolic acid content (F1PA1, F1PA2, F1PA3) and other antinutritive substances. Marker-assisted selection with reference to this locus could achieve substantial reductions in phenolic acid and fibre components in canola meal, independent of seed colour.
The QTL on linkage groups C01 and C05 for phenolic acids (F1PA1, F1PA2), seed colour and other antinutritional traits coincide with the positions of two copies of the gene BnSGT1-(sinapoylglucose transferase) in the YE2-DH population (Lipsa et al., 2012). SGT1 is a major gene involved in sinapate ester biosynthesis and is expressed during seed formation. The obtained results suggest a possible interaction between Bntt1 and BnSGT1 in the regulation of phenylpropanoid biosynthesis in the seed coat.
Today, to identify more closely linked molecular markers than the reported SSR markers, advanced resources are available, including a single nucleotide polymorphism (SNP) map for the YE2 population (Stein et al., 2013) that could be used to assign QTL by comparison to the new reference genome from Express 617 (Lee et al., 2020). This information could be used to more closely identify sequence-annotated markers flanking the QTL and possibly narrow down the candidate genes.
CONCLUSIONS
Two distinct peaks of phenolic acids were detected during the HPLC characterisation of crude phenolic extracts. These are thought to represent the phenolic acids sinapine (F1PA1) and sinapoylglucose (F1PA2), the two most common in rapeseed meal. F1PA1 and F1PA2 QTLs, as well as a seed colour QTL, were clustered together on chromosome C01. The primary QTL for seed colour (part. R2=40.9%) and additional QTL for total flavonoid compounds, ADF, NDF and ADL content were all located on the same region of chromosome A09. This region also contained two loci for F1PA2 (probably sinapoylglucose) and F1PA3. Because a correlation was observed between seed coat thickness and amounts of phenolic acids and other antinutritive chemicals, this research has important implications for breeding to improve the quality of rapeseed meal. More precise mapping of the major QTL using dense, sequence-annotated SNP markers, for example, from a denser SNP map of the YE2 population published by Stein et al. (2013), may help identify sequence-annotated markers flanking the QTL. Using closely linked SNPs for MAS could significantly reduce antinutritional phenolic acids and fibre components in rapeseed meal, independent of seed colour.
Author Contributions: conceptualization, W.F., R.S.; methodology, F.D., R.S.; investigation, F.D.L..; formal analisys, F.D.L.; data curation, F.D.L.; writing-original draft preparation, F.D.L; writing-review and editing, F.D.L. B.W.; supervision, W.F., R.S. All authors have read and agreed to the published version of the manuscript.
Funding: This work was performed with funding from the Federal Ministry of Education and Research (BMBF) as part of the collaborative research project GABI-CGAT: YelLowSin with support from the commercial partners KWS Saat AG, Deutsche Saatveredelung AG, Norddeutsche Pflanzenzucht Hans-Georg Lembke KG and SAATEN-UNION BIOTEC GmbH.
Acknowledgements. The first author would like to gratefully acknowledge financial support from the German Academic Exchange Service (DAAD).
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
Aludatt, M.H.; Rababah, T.; Alhamad, M.N.; Al-Mahasneh, M.A.; Almajwal, A.; Gammoh, S.; Alli, I. A review of phenolic compounds in oil-bearing plants: Distribution, identification and occurrence of phenolic compounds. Food chemistry. 2017, 218, 99-106. https://doi.org/10.1016/j.foodchem.2016.09.057.
Bekele, W.A. Cloning and characterization of a candidate gene for seed coat lignin biosynthesis in oilseed rape. Master Thesis, University Justus-Liebig Giessen, 2008.
Cheng, H.; Liu, X.; Xiao, Q.; Zhang, F.; Liu, N.; Tang, L.; Wang, J.; Ma, X.; Tan, B.; Chen, J.; Jiang, X. Rapeseed meal and its application in pig diet: a review. Agriculture. 2022, 12, 849. https://doi.org/10.3390/agriculture12060849.
Corso, M.; Perreau, F.; Mouille, G.; Lepiniec, L. Specialized phenolic compounds in seeds: Structures, functions, and regulations. Plant Science. 2020, 296, 110471. https://doi.org/10.1016/j.plantsci.2020.110471.
Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus. 1990, 12, 13-15.
Gacek, K.; Bayer, P.E.; Anderson, R.; Severn-Ellis, A.A.; Wolko, J.; Łopatyńska, A.; Matuszczak, M.; Bocianowski, J.; Edwards, D.; Batley, J. QTL genetic mapping study for traits affecting meal quality in winter oilseed rape (Brassica napus L.). Genes. 2021, 12, 1235. https://doi.org/10.3390/genes12081235
Kolodziejczyk, P.; Xiaoyan, W.; Marianchuk, M.; Wanli, L.; Amarowicz, R. Phenolics in Rapeseed: Capillary Electrophoresis as a Novel Analytical Method for Detection of Sinapine, Sinapic Acid Esters and Ferulates. Proceedings of the 10th International Rapeseed Congress, Canberra, Australia. Vol. I, pp. 478-491.
Kozlowska, H.; Naczk, M.; Shahidi, F.; Zadernowski, R. Canola and Rapeseed. Production, Chemistry, Nutrition and Processing Technology. Van Nostrand Reinhold, New York, USA, 1990, pp. 193-210.
Krygier, K.; Sosulski, F.; Hogge, L. Free, esterified, and insoluble-bound phenolic acids. 2. Composition of phenolic acids in rapeseed flour and hulls. Journal of Agricultural and Food Chemistry. 1982, 30, 334-336. https://doi.org/10.1021/jf00110a029.
Lee, H.; Chawla, H.S.; Obermeier, C.; Dreyer, F.; Abbadi, A.; Snowdon, R. Chromosome-scale assembly of winter oilseed rape Brassica napus. Frontiers in plant science, 2020, 11, 496. https://doi.org/10.3389/fpls.2020.00496.
Lipsa, F.D.; Snowdon, R.; Friedt, W. Quantitative genetic analysis of condensed tannins in oilseed rape meal. Euphytica. 2012, 184, 195-205. https://doi.org/10.1007/s10681-011-0546-3.
Mabon, N.; Wathelet, J.P.; Marlier, M. Measurement of Aromatic Choline Esters in Rapeseed by HPLC. Proceedings of the 10th International Rapeseed Congress, Canberra, Australia. Vol. I, pp. 118-122.
Milkowski, C.; Baumert, A.; Schmidt, D.; Nehlin, L.; Strack, D. Molecular regulation of sinapate ester metabolism in Brassica napus: expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant Journal. 2004, 38, 80-92. https://doi.org/10.1111/j.1365-313X.2004.02036.x.
Milkowski, C.; Strack, D. Sinapate esters in brassicaceous plants: biochemistry, molecular biology, evolution and metabolic engineering. Planta. 2010, 232, 19-35. https://doi.org/10.1007/s00425-010-1168-z.
Naczk, M.; Amarowicz, R.; Sullivan, A.; Shahidi, F. Current research developments on polyphenolics of rapeseed/canola: a review. Food Chemistry. 1998, 62, 489-502. https://doi.org/10.1016/S0308-8146(97)00198-2.
Naczk, M.; Shahidi, F. Extraction and analysis of phenolics in food. Journal of Chromatography A. 2004, 1054, 95-111. https://doi.org/10.1016/j.chroma.2004.08.059.
Obied H.K.; Song Y.; Foley S.; Loughlin M.; Rehman A.; Mailer R.; Masud T.; Agboola S. Biophenols and antioxidant properties of Australian canola meal. Journal of Agricultural and Food Chemistry. 2013, 61, 9176-9184. https://doi.org/10.1021/jf4026585.
Qiao, H.; Classen, H.L. Nutritional and physiological effects of rapeseed meal sinapine in broiler chickens and its metabolism in the digestive tract. Journal of the Science of Food and Agriculture. 2003, 83, 1430-1438. https://doi.org/10.1002/jsfa.1559.
Reungoat, V.; Allais, F.; Ducatel, H.; Ioannou, I. Extraction and purification processes of sinapic acid derivatives from rapeseed and mustard seed by-products. Separation & Purification Reviews. 2022, 51, 521-544. https://doi.org/10.1080/15422119.2021.2004550.
Rezaeizad, A.; Wittkop, B.; Snowdon, R.; Hasan, M.; Mohammadi, V.; Zali, A.; Friedt, W. Identification of QTLs for phenolic compounds in oilseed rape (Brassica napus L.) by association mapping using SSR markers. Euphytica. 2011, 177, 335-342. https://doi.org/10.1007/s10681-010-0231-y.
Shahidi, F.; Naczk, M. Methods of analysis and quantification of phenolic compounds. Chemistry, Effects, and Applications. Technomic Publication Co. Inc., Lancaster, USA, 1995, pp 20-65.
Stein, A.; Wittkop, B.; Liu, L.; Obermeier, C.; Friedt, W.; Snowdon, R.J. Dissection of a major QTL for seed colour and fibre content in Brassica napus reveals colocalization with candidate genes for phenylpropanoid biosynthesis and flavonoid deposition. Plant Breeding. 2013, 132, 382-389. https://doi.org/10.1111/pbr.12073.
Sun, B.S.; Leandro, M.C.; De Freitas, V.; Spranger, M.I. Fractionation of red wine polyphenols by solid-phase extraction and liquid chromatography, Journal of Chromatography A. 2006, 1128, 27-38. https://doi.org/10.1016/j.chroma.2006.06.026.
Tan, S.H.; Mailer, R.J.; Blanchard, C.L.; Agboola, S.O. Canola Proteins for Human Consumption: Extraction, Profile, and Functional Properties. Journal of Food Science, 2011, 76, 16-28. https://doi.org/10.1111/j.1750-3841.2010.01930.x.
TUM (Technical University of Munich). How rapeseed could be used as a protein source for human nutrition. ScienceDaily. 2019. www.sciencedaily.com/releases/2019/01/190131125936.htm (accessed on 10 December 2022).
Vuorela, S.; Meyer, A.S.; Heinonen, M. Quantitative analysis of the main phenolics in rapeseed meal and oils processed differently using enzymatic hydrolysis and HPLC. European Food Research and Technology. 2003, 217, 517-523. https://doi.org/10.1007/s00217-003-0811-3.
Wittkop, B.; Snowdon, R.J.; Friedt, W. Status and perspectives of breeding for enhanced yield and quality of oilseed crops for Europe. Euphytica. 2009, 170, 131-140. https://doi.org/10.1007/s10681-009-9940-5.
Yang, S.; Arasu, M.; Chun, J.; Jang, Y.; Lee, Y.; Kim, I.; Lee, K.; Hong, S.; Kim, S. Identification and determination of phenolic compounds in rapeseed meals (Brassica napus L.). Journal of Agricultural Chemistry and Environment. 2015, 4, 14-23. https://doi.org/10.4236/jacen.2015.41002.
Yang, Z.; Huang, Z.; Cao, L. Biotransformation technology and high-value application of rapeseed meal: a review. Bioresources and Bioprocessing. 2022, 9, 1-18. https://doi.org/10.1186/s40643-022-00586-4.
Zum Felde, T.; Baumert, A.; Strack, D.; Becker, H.C.; Möllers, C. Genetic variation for sinapate ester content in winter rapeseed (Brassica napus L.) and development of NIRS calibration equation. Plant Breeding. 2007, 126, 291-296. https://doi.org/10.1111/j.1439-0523.2007.01342.x.
*Brassica. http://www.brassica.info/resource/maps/lg-assignments.php (accessed on 10 November 2022).
Academic Editor: Prof. Dr. Maria Harja
Publisher Note: Regarding jurisdictional assertions in published maps and institutional affiliations ALSE maintain neutrality.
Friedt Wolfgang, Lipșa Florin Daniel, Snowdon Rod, Wittkop Benjamin



