Andreea Paula Cozma, Iulia Elena Măciucă, Cristina Mihaela Rîmbu, Ioana Crivei, Șerban Moroșan, Lucia Carmen Trincă, Dorina Timofte
ABSTRACT. Antimicrobial resistance is a major public health concern worldwide. This study aims to determine the prevalence of Enterobacterales producing beta-lactamase (TEM, SHV, OXA) or extended-spectrum beta-lactamases (ESBL), as well as plasmid-mediated resistance to quinolones (PMQR) (qnrA, qnrB, qnrS) in companion animals from the northeast region of Romania. A total of 124 faecal samples were collected aseptically from healthy dogs attending the veterinary practice for vaccination and cultivated on Brilliance ESBL medium (Oxoid, UK). The ESBL production testing was performed using the combination disc test. The identification of Enterobacterales strains was achieved using molecular identification and based on biochemical tests. Antimicrobial susceptibility testing was performed using the disk diffusion method. Identification of genes encoding for beta-lactamase enzymes and genes encoding plasmid-mediated resistance to quinolones was performed by PCR according to the protocols previously described. After ESBL screening, 31 (31/124; 25%) extended-spectrum cephalosporin (ESC)-resistant Enterobacterales were obtained, and 67.74% (21/31) of them were confirmed as ESBL-producers. Regarding the Enterobacterales species, 27 (27/31; 87.1%) were Escherichia coli and 4 (4/31; 12.9%) strains were Klebsiella pneumoniae. Among the ESBL-producing isolates, the blaCTX-M-1 gene group was predominant (58.82%), followed by the blaCTX-M-9 group (41.18%). The blaTEM, blaSHV and blaOXA gene groups were identified in 54.83%, 29.03% and 3.22% of the analysed strains, respectively. The prevalence of PMQR genes was 22.58% and consisted only of qnrS (19.35%) and qnrA (3.22%) genes. The prevalence of ESBL strains related to the total number of analysed samples was 16.93% (21/124). The findings show a significant prevalence of ESBLs and PMQR genes in Enterobacterales strains isolated from the faeces of healthy dogs, implying that pets may pose a risk of transmitting ESBL strains to other animals or owners.
Keywords: antimicrobial resistance; companion animals; ESBL genes.
Cite
ALSE and ACS Style
Cozma, A.P.; Măciucă, I.E.; Rîmbu, C.M.; Crivei, I.; Moroșan, Ș.; Trincă, L.C.; Timofte, D. Prevalence and characterisation of extended-spectrum beta-lactamases and plasmid-mediated quinolones resistance in Enterobacteriaceae isolated from companion animals. Journal of Applied Life Sciences and Environment 2023, 56 (4), 541-549.
https://doi.org/10.46909/alse-564115
AMA Style
Cozma AP, Măciucă IE, Rîmbu CM, Crivei I, Moroșan Ș, Trincă LC, Timofte D. Prevalence and characterisation of extended-spectrum beta-lactamases and plasmid-mediated quinolones resistance in Enterobacteriaceae isolated from companion animals. Journal of Applied Life Sciences and Environment. 2023; 56 (4), 541-549.
https://doi.org/10.46909/alse-564115
Chicago/Turabian Style
Cozma, Andreea Paula, Iulia Elena Măciucă, Cristina Mihaela Rîmbu, Ioana Crivei, Șerban Moroșan, Lucia Carmen Trincă, and Dorina Timofte. 2023. “Prevalence and characterisation of extended-spectrum beta-lactamases and plasmid-mediated quinolones resistance in Enterobacteriaceae isolated from companion animals” Journal of Applied Life Sciences and Environment 56, no. 4: 541-549.
https://doi.org/10.46909/alse-564115
View full article (HTML)
Prevalence and Characterisation of Extended-Spectrum Beta-Lactamases and Plasmid-Mediated Quinolones Resistance in Enterobacteriaceae Isolated from Companion Animals
Andreea Paula COZMA1*, Iulia Elena MĂCIUCĂ2, Cristina Mihaela RÎMBU3, Ioana CRIVEI3, Șerban MOROȘAN3,4, Lucia Carmen TRINCĂ1 and Dorina TIMOFTE3,2
1Department of Exact Sciences, Faculty of Horticulture, “Ion Ionescu de la Brad” Iasi University of Life Sciences, 3, Mihail Sadoveanu Alley, 700490, Iasi, Romania; email: lctrinca@uaiasi.ro
2Department of Veterinary Anatomy, Physiology and Pathology, School of Veterinary Science, Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Neston, United Kingdom; e-mail: iuliana.maciuca@liverpool.ac.uk; d.timofte@liverpool.ac.uk
3Department of Public Health, Faculty of Veterinary Medicine, “Ion Ionescu de la Brad” Iasi University of Life Sciences, 8, Mihail Sadoveanu Alley, 700489, Iasi, Romania; email: crimbu@yahoo.com; ioana.crivei@yahoo.com
4Sorbonne University, UMS 28 INSERM, Paris, France; e-mail: serban.morosan@sorbonne-universite.fr
*Correspondence: andreeapaulacozma@yahoo.com
Received: Oct. 30, 2023. Revised: Dec. 09, 2023. Accepted: Dec. 14, 2023. Published online: Jan. 15, 2024
ABSTRACT. Antimicrobial resistance is a major public health concern worldwide. This study aims to determine the prevalence of Enterobacterales producing beta-lactamase (TEM, SHV, OXA) or extended-spectrum beta-lactamases (ESBL), as well as plasmid-mediated resistance to quinolones (PMQR) (qnrA, qnrB, qnrS) in companion animals from the northeast region of Romania. A total of 124 faecal samples were collected aseptically from healthy dogs attending the veterinary practice for vaccination and cultivated on Brilliance ESBL medium (Oxoid, UK). The ESBL production testing was performed using the combination disc test. The identification of Enterobacterales strains was achieved using molecular identification and based on biochemical tests. Antimicrobial susceptibility testing was performed using the disk diffusion method. Identification of genes encoding for beta-lactamase enzymes and genes encoding plasmid-mediated resistance to quinolones was performed by PCR according to the protocols previously described. After ESBL screening, 31 (31/124; 25%) extended-spectrum cephalosporin (ESC)-resistant Enterobacterales were obtained, and 67.74% (21/31) of them were confirmed as ESBL-producers. Regarding the Enterobacterales species, 27 (27/31; 87.1%) were Escherichia coli and 4 (4/31; 12.9%) strains were Klebsiella pneumoniae. Among the ESBL-producing isolates, the blaCTX-M-1 gene group was predominant (58.82%), followed by the blaCTX-M-9 group (41.18%). The blaTEM, blaSHV and blaOXA gene groups were identified in 54.83%, 29.03% and 3.22% of the analysed strains, respectively. The prevalence of PMQR genes was 22.58% and consisted only of qnrS (19.35%) and qnrA (3.22%) genes. The prevalence of ESBL strains related to the total number of analysed samples was 16.93% (21/124). The findings show a significant prevalence of ESBLs and PMQR genes in Enterobacterales strains isolated from the faeces of healthy dogs, implying that pets may pose a risk of transmitting ESBL strains to other animals or owners.
Keywords: antimicrobial resistance; companion animals; ESBL genes.
INTRODUCTION
Antimicrobial resistance (AMR) is a major public health issue affecting both human and veterinary medicine. Extended-spectrum beta-lactamase (ESBL) enzymes provide resistance to third and fourth-generation cephalosporins and also to aztreonam, the newest antibiotic available to treat enterobacteria infections, such as Escherichia coli or Klebsiella pneumoniae (Bush and Jacoby, 2010). Research on extended-spectrum cephalosporin (ESC)-resistant Enterobacteriaceae strains isolated from pets has increased over the past 20 years.
However, the prevalence of these strains in some countries, especially in less developed countries, has not been reported. Furthermore, although antibiotic use in domestic animals is widespread, relatively few studies have quantified antibiotic usage and AMR in dogs and cats compared to the research in other species or categories of animals and humans. It is known that very close daily contact with humans is an important risk factor for transmission of these strains within or between species.
The aim of this study was to determine the prevalence of Enterobacteriaceae strains that produce beta-lactamase enzymes (TEM, SHV, OXA), ESBL, and genes encoding plasmid-mediated resistance to quinolones (PMQR) (qnrA, qnrB, qnrS).
MATERIALS AND METHODS
Faecal samples were collected aseptically using rectal swabs from clinically healthy dogs that came to the veterinary clinic for vaccination. After collection, the samples were immediately inoculated onto the screening medium Brilliance ESBL (Oxoid, Basingstoke, UK), as described in a previous article by the authors (Cozma et al., 2019).
The bacterial strains that showed characteristic Enterobacteriaceae colonies on the screening medium were subcultured onto blood agar medium (Oxoid, Basingstoke, UK) for subsequent testing and identification. For E. coli isolates, species confirmation was performed by PCR based on molecular identification of the uidA and uspA genes (Anastasi et al., 2010; McDaniels et al., 1996) and for the K. pneumoniae isolates, based on biochemical tests (API 20E, Biomerieux, Marcy-l’Étoile, France).
Phenotypic characterisation of ESBL production was performed for all isolated strains (n = 31). The colonies identified as E. coli and K. pneumoniae were subcultured onto blood agar medium and tested using the combination disc test (Cozma et al., 2019). All isolates (n = 31) were also antibiotic susceptibility tested using the disk diffusion method on Muller–Hinton agar medium.
The data were interpreted in conformity with the Clinical and Laboratory Standards Institute (CLSI) recommendations (CLSI, 2018). If an isolate had intermediate or resistant results against the tested antimicrobial agent, it was considered non-susceptible.
The following antibiotics were included in the antimicrobial panel: ampicillin (10μg), amoxicillin/clavulanic acid (30μg), imipenem (10μg), aztreonam (30μg), enrofloxacin (5μg), trimethoprim/ sulfamethoxazole (25μg), tetracycline (30μg), chloramphenicol (30μg), and gentamicin (10μg). One ATCC standard strain of E. coli (E. coli ATCC 25,922) was used as a control strain.
Genetic background characterisation was done by PCR for the genes that produce beta-lactamase enzymes, ESBL and PMQR. The extraction of bacterial DNA was performed using the boiled preps method (Maciuca et al., 2015). By using PCR according to the previously described protocols, we aimed to identify the gene groups blaCTX-M, blaTEM, blaSHV and the PMQR genes, respectively (Dallenne et al., 2010; Robicsek et al., 2006; Wedley et al., 2011)
RESULTS
A total of 124 faecal samples were collected aseptically from clinically healthy dogs that came to the veterinary clinic for vaccination. Following ESBL screening, 31 (31/124; 25%) Enterobacteriaceae strains resistant to extended cephalosporins were obtained, of which 27 (27/31; 87.1%) were E. coli, and 4 (4/31; 12.9%) strains were K. pneumoniae.
All 31 extended-spectrum cephalosporin-resistant (ESC-R) Enterobacterales were tested for antimicrobial susceptibility. All organisms were resistant to ampicillin, with 77.41% resistant to amoxicillin/ clavulanic acid, 61.29% resistant to sulfamethoxazole/trimethoprim, 58.06% resistant to tetracycline, and 45.16% resistant to chloramphenicol, gentamicin and enrofloxacin (Figure 1).
All isolated strains were analysed to assess the degree of multidrug resistance (MDR). According to Magiorakos et al. (2012), resistance to more than three classes of antibiotics defines a strain as being MDR. Following the analysis of the results obtained in the disk diffusion antibiotic susceptibility test, 18 (18/31; 58.06%) strains were associated with MDR.
Molecular characterisation of the genetic background was done for the 31 ESC-R Enterobacterales carrying genes or gene combinations (Table 1). The most prevalent groups of genes were the blaCTX-M gene group (54.83%; 17/31) and the blaTEM gene group (54.83%; 17/31) (Figure 2). In the blaCTX-M gene group, the blaCTX-M-1 group was identified as predominant (10/17; 58.82%), along with the blaCTX-M-9 group (7/17; 41.18%) (Figure 2).
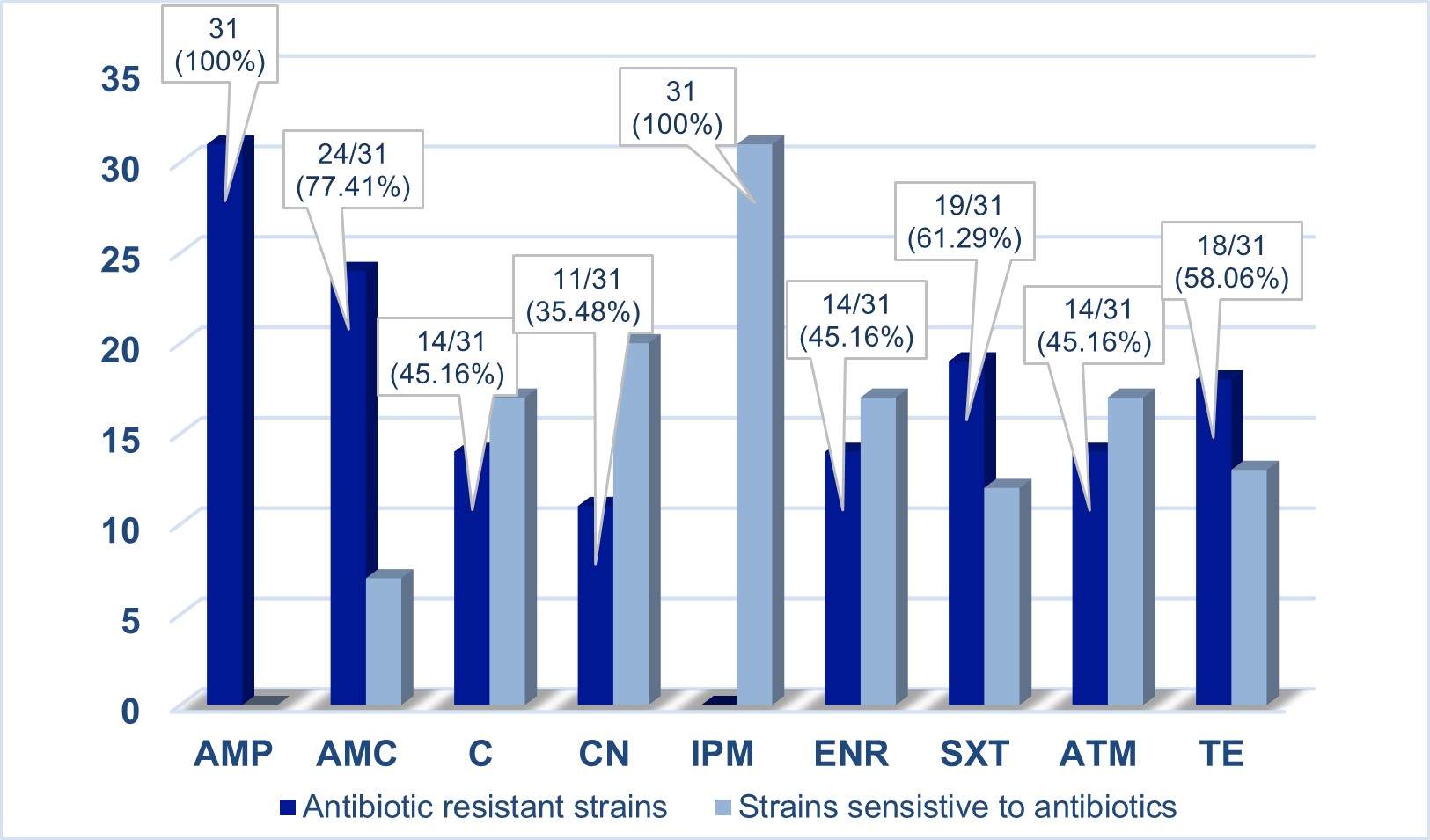
Figure 1 – Antimicrobial susceptibility testing results (n = 31 isolates) Abbreviations: AMP, ampicillin; AMC, amoxicillin/clavulanic acid; C, chloramphenicol; CN, gentamicin; IPM, imipenem; ENR, enrofloxacin; SXT, trimethoprim/sulfamethoxazole; ATM, aztreonam; TE, tetracycline
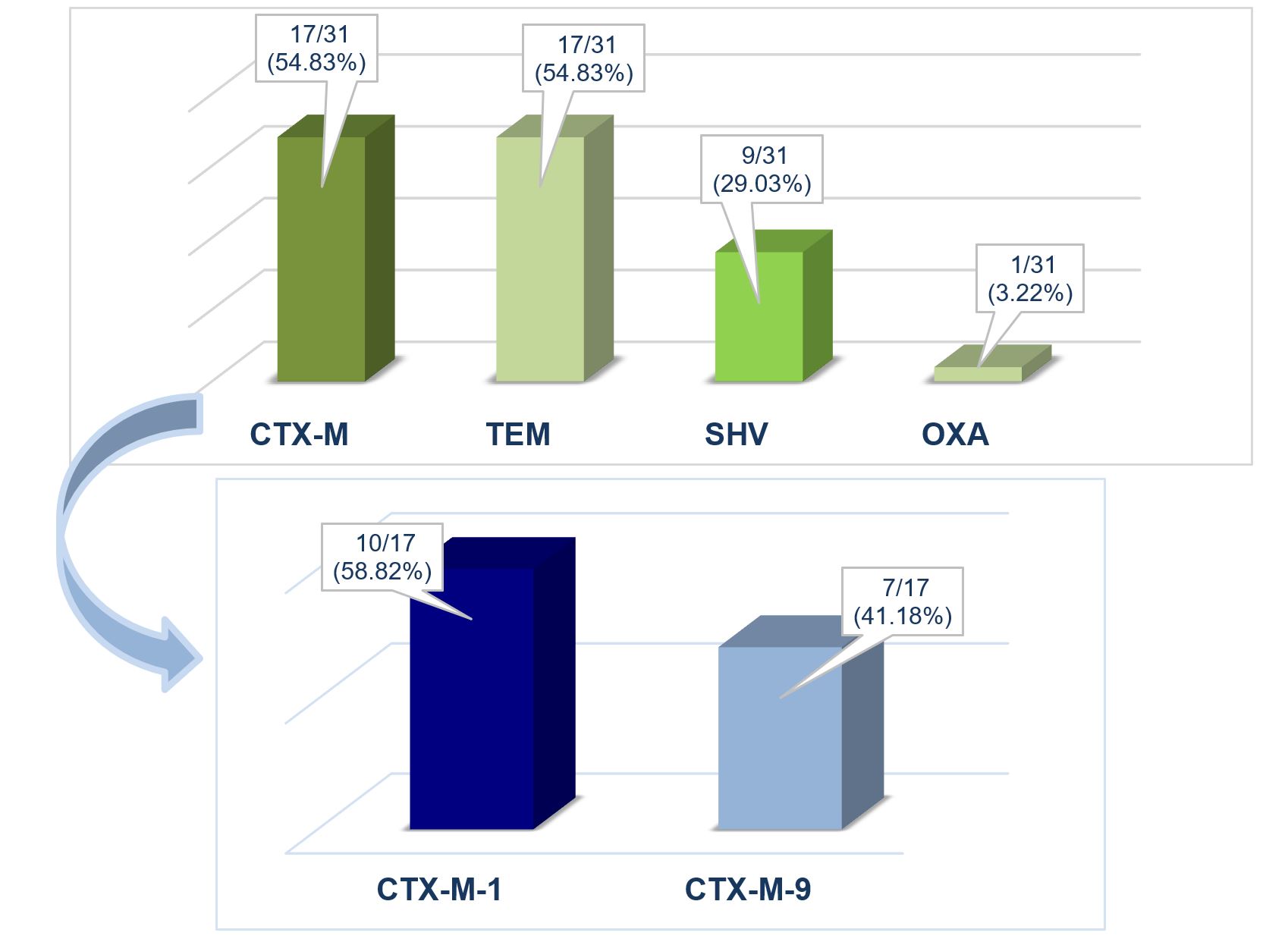
Figure 2 – Molecular characterisation of the genetic background regarding antibiotic resistance of the isolated strains
The blaSHV and blaOXA gene groups were identified in 9/31 (29.03%) and 1/31 (3.22%), respectively, of the analysed strains (Figure 2). In some isolates, only the blaTEM gene (6/31; 19.35%), the blaSHV gene (4/31; 12.9%), or combinations of the blaTEM and blaSHV (3/31; 9.68%) were present, and only one strain (1/31; 3.22%) had the genes blaTEM, blaSHV and blaOXA (Table 1). The prevalence of PMQR genes was 22.58% (7/31), and only the qnrS (6/31; 19.35%) and qnrA (1/3; 3.22%) genes were identified (Figure 3).
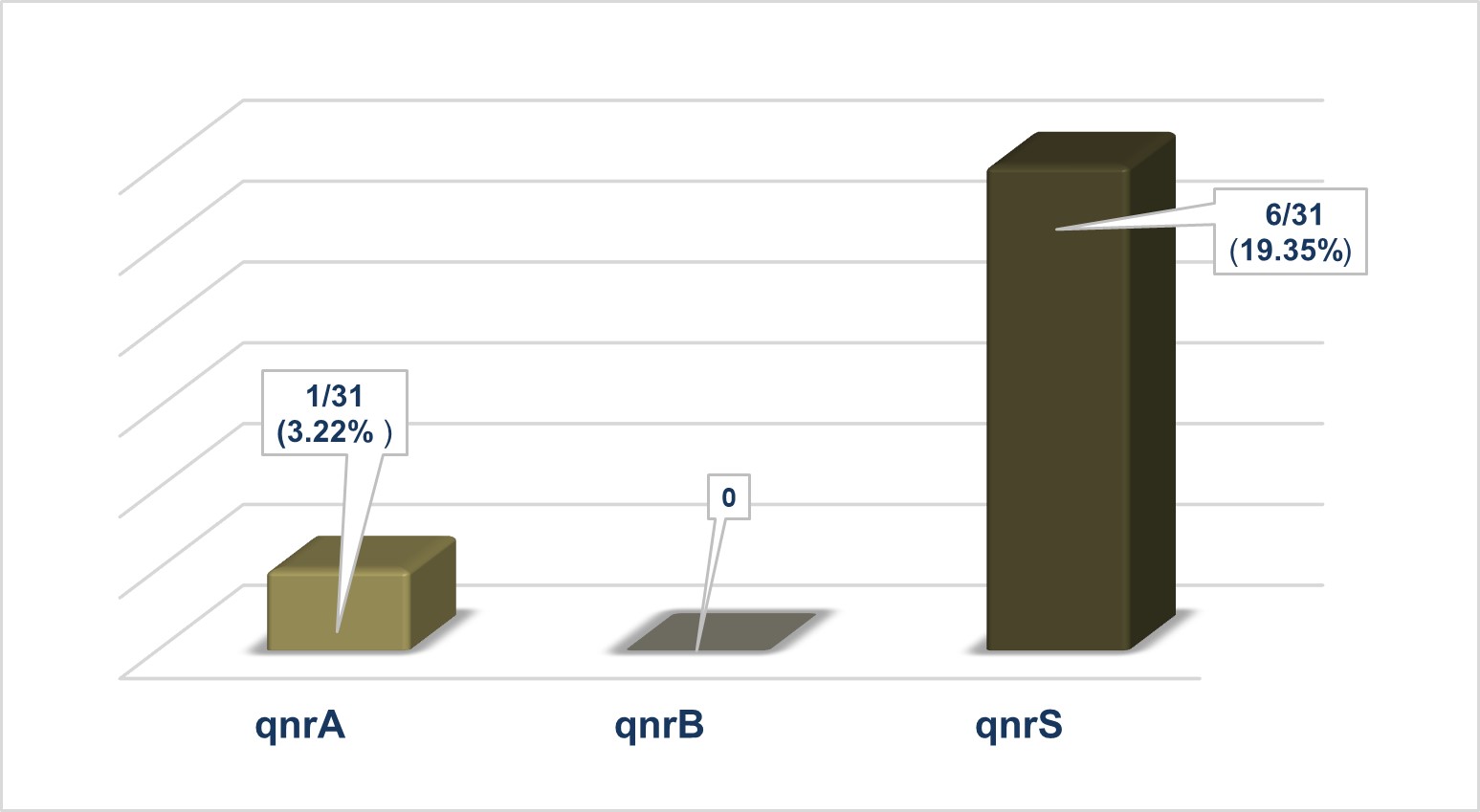
Figure 3 – Molecular characterisation of the genetic background regarding plasmid-mediated resistance to quinolones
Table 1
Molecular identification of extended-spectrum cephalosporin-resistant (ESC-R) Enterobacterales in dog faeces
|
Bacterial species |
No. and isolate ID |
ESBL phenotype YES/NO |
Beta-lactamase genes by PCR |
Associated PMQR genes |
|
E. coli |
3 (MV67;MV46;MV67i) |
YES |
blaCTX-M-1 group |
– |
|
5 (MV18;MV19;MV22;MV21;MV40) |
YES |
blaCTX-M-1 group |
qnrS |
|
|
1 (MV73) |
YES |
blaCTX-M-1 group; blaTEM |
qnrS |
|
|
1 (MV47) |
YES |
blaCTX-M-1 group; blaTEM; blaSHV |
– |
|
|
2 (MV39;MV43) |
YES |
blaCTX-M-9 group |
– |
|
|
5 (MV20;MV66;MV16;MV66p;MV17) |
YES |
blaCTX-M-9 group; blaTEM |
– |
|
|
1 (MV37) |
YES |
blaTEM |
– |
|
|
5 (MV29; MV30; MV33; MV62; MV63) |
NO |
blaTEM |
– |
|
|
1 (MV38) |
YES |
blaTEM; blaSHV |
– |
|
|
1 (MV34) |
YES |
blaTEM; blaSHV |
qnrA |
|
|
1 (MV31) |
YES |
blaTEM; blaSHV; blaOXA |
– |
|
|
1 (MV71) |
NO |
blaTEM; blaSHV; |
– |
|
|
K. pneumoniae |
14 (MV71p; MV71K; MV68E; MV68) |
NO |
blaSHV |
– |
The strains carrying blaCTX-M genes are considered ESBL enzyme-producing strains (Zeynudin et al., 2018). The blaTEM, blaSHV or blaOXA gene groups encode both beta-lactamase and ESBL enzymes (Ewers et al., 2011). In this study, the strains that were carriers of the blaTEM, blaSHV or blaOXA genes were confirmed as being ESBL-producing strains correlating the molecular results with the results obtained in the combination disc method.
Therefore, the prevalence of ESBL strains, of the 31 strains resistant to extended cephalosporins, was 67.74% (21/31). The prevalence of ESBL strains related to the total number of analysed samples was 16.93% (21/124).
DISCUSSION
In Romania, there are more publications on the prevalence of AMR in human bacterial isolates (ECDC, 2022), but for veterinary medicine, the data are limited. This study has analysed bacterial phenotypes in combination with the genetic characteristics of E. coli and K. pneumoniae strains isolated from healthy dogs from the northeast region of Romania.
The 16.93% prevalence of ESBL strains in this study is much higher than the global average (6.87%) identified in dogs (Salgado-Caxito et al., 2021) and similar to the prevalence obtained in strains isolated from chicken in Romania (Maciuca et al., 2015). Also, compared to other similar studies carried out in Europe, the obtained prevalence is lower than in countries such as France (18.5%) or Spain (19.6%) (Abreu-Salinas et al., 2020; Haenni et al., 2014).
The blaCTX-M genotype was most commonly identified among the isolated strains and was reported in approximately 95% of similarly conducted studies (Salgado-Caxito et al., 2021). Moreover, the blaCTX-M-1 genotype is the most commonly identified in similar studies. In addition, like in similar studies, our study showed that the qnr gene family, qnrs, was the most prevalent. Moreover, the PMQR variants (qnrS and qnrA) were coexpressed with the beta-lactamase or ESBL enzymes (Cui et al., 2022; de Jong et al., 2018).
The World Health Organization (WHO) has stated that quinolone antibiotics, beta-lactam antibiotics (such as third, fourth and fifth-generation cephalosporins, and aminopenicillins with and without beta-lactamase inhibitors) and aminoglycosides are antibiotics of critical importance (WHO, 2007).
All of the above-mentioned antibiotics are also used in veterinary medicine and this study has shown increased resistance to some of the tested antibiotics of critical importance.
The percentage of antibiotic-resistant strains varies by country. Antibiotic resistance is also highly dependent on the implementation of public policies regulating antibiotic prescribing, especially in veterinary medicine, which has been shown to be related to the emergence of MDR strains.
Moreover, the use of antibiotics in animal feed, without quantification or with empirical dosing, and the preferential use of enrofloxacin and doxycycline without alternating them with other antibiotics were other factors influencing the emergence of MDR strains in veterinary medicine (Dierikx et al., 2012).
CONCLUSIONS
Researchers discovered a high incidence of ESBLs in Enterobacteriaceae strains isolated from the faeces of clinically healthy companion animals (dogs), indicating the risk of ESBL strains spreading to other animals or owners.
Research on ESC-resistant Enterobacteriaceae in companion animals has increased in recent years, showing that these bacteria are present in dogs worldwide. However, their prevalence in some companion animal populations from many countries, including Romania, has not yet been reported; this is concerning given the shared environment and close contact with the owners.
Future research should focus on the identification of the factors responsible for the acquisition and dissemination of ESC-resistant Enterobacteriaceae in pets, including interspecies transmission, clinical relevance and their economic impact.
Author Contributions: Conceptualization, D.T., A.P.C; methodology, D.T., A.P.C., and I.E.M.; validation, D.T., A.P.C.; C.M.R., M.S. AND L.C.T.; formal analysis, A.P.C., C.M.R., I.E.M.; investigation, A.P.C., I.E.M. and I.C.; resources, D.T.; data curation, A.P.C., I.C. and C.M.R.; writing—original draft preparation, A.P.C.; writing—review and editing, D.T. and A.P.C., C.M.R. and I.E.M.; supervision, D.T.; L.C.T.; M.S.; project administration, D.T.; funding acquisition, D.T. All authors have read and agreed to the published version of the manuscript.
Funding: There was no external funding for this study.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
Abreu-Salinas, F.; Díaz-Jiménez, D.; García-Meniño, I.; Lumbreras, P.; López-Beceiro, A.M.; Fidalgo, L.E.; Fernández, J. High prevalence and diversity of cephalosporin-resistant Enterobacteriaceae including extraintestinal pathogenic E. coli CC648 lineage in rural and urban dogs in Northwest Spain. Antibiotics. 2020, 9, 468. https://doi.org/10.3390/antibiotics9080468.
Anastasi, E.M.; Matthews, B.; Gundogdu, A.; Vollmerhausen, T.L.; Ramos, N.L.; Stratton, H.; Ahmed, W.; Katouli, M. Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Applied and Environmental Microbiology. 2010, 76, 5882-5886. https://doi.org/10.1128/aem.00141-10.
Bastidas-Caldes, C.; Romero-Alvarez, D.; Valdez-Vélez, V.; Morales, R.D.; Montalvo-Hernández, A.; Gomes-Dias, C.; Calvopiña, M. Extended-spectrum beta-lactamases producing Escherichia coli in South America: a systematic review with a One Health perspective. Infection and drug resistance. 2022, 5759-5779. https://doi.org/10.2147/idr.s371845.
Bush, K.; Jacoby, G.A. Updated functional classification of β-lactamases. Antimicrobial agents and chemotherapy. 2010, 54, 969-976. https://doi.org/10.1128/aac.01009-09.
CLSI (Approved Standard M100-S15). Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
Cozma, A.P.; Maciuca, I.E.; Carp-Cărare, C.; Carp-Cărare, M.; Rîmbu, C.; Aniță, A.; Aniță, D.; Timofte, D. Characterisation of Extended β-Lactamases and Plasmid Mediated Quinolones Resistance in Escherichia coli from Shelter Dogs. Bulletin UASVM Veterinary Medicine. 2019, 76, 100-103.
Cui, L.; Zhao, X.; Li, R.; Han, Y.; Hao, G.; Wang, G.; Sun, S. Companion Animals as Potential Reservoirs of Antibiotic Resistant Diarrheagenic Escherichia coli in Shandong, China. Antibiotics. 2022, 11, 828. https://doi.org/10.3390/antibiotics11060828.
Dallenne, C.; Da Costa, A.; Decre, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy. 2010, 65, 490-495. https://doi.org/10.1093/jac/dkp498.
de Jong, A.; Muggeo, A.; El Garch, F.; Moyaert, H.; de Champs, C.; Guillard, T. Characterization of quinolone resistance mechanisms in Enterobacteriaceae isolated from companion animals in Europe (ComPath II study). Veterinary microbiology. 2018, 216, 159-167. https://doi.org/10.1016/j.vetmic.2018.02.002.
Dierikx, C.M.; van Duijkeren, E.; Schoormans, A.H.W.; van Essen-Zandbergen, A.; Veldman, K.; Kant, A.; Mevius, D.J. Occurrence and characteristics of extended-spectrum-β-lactamase-and AmpC-producing clinical isolates derived from companion animals and horses. Journal of Antimicrobial Chemotherapy. 2012, 67, 1368-1374.
ECDC (European Centre for Disease Prevention and Control). Antimicrobial Resistance Surveillance in Europe 2022–2020 Data; World Health Organization, Regional Office for Europe: Copenhagen, Denmark, 2022.
Ewers, C.; Grobbel, M.; Bethe, A; Wieller, L.H.; Guenther S. Extended-spectrum beta-lactamases-producing Gram-negative bacteria in companion animals: action is clearly warranted!. Berlin Und Munchener Tierarztliche Wochenschrift. 2011, 124, 94-101.
Haenni, M.; Saras, E.; Métayer, V.; Médaille, C.; Madec, J.Y. High prevalence of blaCTX-M-1/IncI1/ST3 and bla CMY-2/IncI1/ST2 plasmids in healthy urban dogs in France. Antimicrobial agents and chemotherapy. 2014, 58, 5358-5362. https://doi.org/10.1128/aac.02545-14.
Maciuca, I.E.; Williams, N.J.; Tuchilus, C.; Dorneanu, O.; Guguianu, E.; Carp-Carare, C.; Timofte, D. High prevalence of Escherichia coli – producing CTX-M-15 extended-spectrum beta-lactamases in poultry and human clinical isolates in Romania. Microbial Drug Resistance. 2015, 21, 651-662. https://doi.org/10.1089/mdr.2014.0248.
Magiorakos, A.-P.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2012, 18, 268-281. https://doi.org/10.1111/j.1469-0691.2011.03570.x.
McDaniels, A.E.; Rice, E.W.; Reyes, A.L.; Johnson, C.H.; Haugland, R.A.; Stelma, G.N. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and beta-D-glucuronidase. Applied and Environmental Microbiology. 1996, 62, 3350-3354. https://doi.org/10.1128/aem.62.9.3350-3354.1996.
Robicsek, A.; Strahilevitz, J.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. qnr Prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrobial Agents and Chemotherapy. 2006, 50, 2872-2874. https://doi.org/10.1128/aac.01647-05.
Salgado-Caxito, M.; Benavides, J.A.; Adell, A.D.; Paes, A.C.; Moreno-Switt, A.I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing- Escherichia coli in dogs and cats– a scoping review and meta-analysis. One Health. 2021, 12, 100236. https://doi.org/10.1016/j.onehlt.2021.100236.
Wedley, A.L.; Maddox, T.W.; Westgarth, C.; Coyne, K.P; Pinchbeck, G.L.; Williams, N.J.; Dawson, S. Prevalence of antimicrobial-resistant Escherichia coli in dogs in a cross-sectional, community-based study. Veterinary Record. 2011, 168, 354. https://doi.org/10.1136/vr.d1540.
WHO. Critically Important Antimicrobials for Human Medicine: Categorization for the Development of Risk Management Strategies to Contain Antimicrobial Resistance Due to Non-Human Antimicrobial Use: Report of the Second WHO Expert Meeting, Copenhagen, 2007. Available online https://apps.who.int/iris/bitstream/handle/10665/43765/9789241595742_eng.pdf
Zeynudin, A.; Pritsch, M.; Schubert, S.; Messerer, M.; Legl, G.; Hoelscher, M.; Belachew, T.; Wieser, A. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infectious Diseases. 2018, 18, 524. https://doi.org/10.1186/s12879-018-3436-7.
Academic Editor: Prof. Dr. Daniel Simeanu
Publisher Note: Regarding jurisdictional assertions in published maps and institutional affiliations ALSE maintain neutrality.
Cozma Andreea Paula, Crivei Ioana Cristina, Măciucă Iulia Elena, Moroșan Șerban, Rîmbu Cristina Mihaela, Timofte Dorina, Trincă Lucia Carmen



