M.H. Shahrajabian, W. Sun, Q. Cheng
ABSTRACT. Chinese medicinal herbs and fruits have grown rapidly and significantly in recent years and have a positive influence on improving people’s attention to their health and organic life style. According to the advancement of sequencing technologies and reduced costs, the genome sequencing data of medicinal plants are accumulating rapidly. Our aim was to review plant genomes of three important medicinal plants in China. There is an ample genetic diversity of plants with medicinal importance around the globe and this pool of genetic variation serves as the base for selection, as well as for plant improvement. Plant genomes are characterized by large variations of genome size and ploidy level. Comparative genomics provides a method to unravel the relationship between genomes, by describing conserved chromosomes or chromosomal regions between related species. It is also clear that it is possible to use plant genome as a tool for improving breeding strategies. However, certain limitations represent a number of challenges for the generation and utilization of genomic resources in many important medicinal plant species. This review has focused on plant genomes of some important horticultural plants, which are famous in traditional Chinese medicine, namely ginger, ginseng and goji berry. However, more researches are needed to introduce the genome research of medicinal plants.
Keywords: gene sequencing; ginger; ginseng; goji berry; horticultural crops; organic life.
View full article (HTML)
A Review on Plant Genomes of Some Important Traditional Chinese Fruits and Herbs
M.H. Shahrajabian1,*, W. Sun1, Q. Cheng1,2 ,*
1Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China
2College of Life Sciences, Hebei Agricultural University, Baoding, Hebei, 071000, China; Global Alliance of HeBAU-CLS&HeQiS for BioAl-Manufacturing, Baoding, Hebei 071000, China
*E-mail: hesamshahrajabian@gmail.com; chengqi@caas.cn
Received: May 10, 2020. Revised: June 15, 2020. Accepted: June 22, 2020. Published online: July 18, 2020
ABSTRACT. Chinese medicinal herbs and fruits have grown rapidly and significantly in recent years and have a positive influence on improving people’s attention to their health and organic life style. According to the advancement of sequencing technologies and reduced costs, the genome sequencing data of medicinal plants are accumulating rapidly. Our aim was to review plant genomes of three important medicinal plants in China. There is an ample genetic diversity of plants with medicinal importance around the globe and this pool of genetic variation serves as the base for selection, as well as for plant improvement. Plant genomes are characterized by large variations of genome size and ploidy level. Comparative genomics provides a method to unravel the relationship between genomes, by describing conserved chromosomes or chromosomal regions between related species. It is also clear that it is possible to use plant genome as a tool for improving breeding strategies. However, certain limitations represent a number of challenges for the generation and utilization of genomic resources in many important medicinal plant species. This review has focused on plant genomes of some important horticultural plants, which are famous in traditional Chinese medicine, namely ginger, ginseng and goji berry. However, more researches are needed to introduce the genome research of medicinal plants.
Keywords: gene sequencing; ginger; ginseng; goji berry; horticultural crops; organic life.
INTRODUCTION
Traditional medicine considers health practices, knowledge, approaches and beliefs incorporating plants, fruits and herbs, based on both ancient and modern pharmaceutical science (Ogbaji et al., 2013, 2018; Soleymani and Shahrajabian 2012, 2018; Shahrajabian et al., 2020a,b,c; Sun et al., 2020a,b). Genome sequencing represents a perfect overview of the structural organization of functional elements in a given genome, and these structural elements carry the knowledge of the evolutionary history of an organism (Xiong et al., 2018; Jiang et al., 2019). Gantait et al. (2014) concluded that there is a plentiful genetic diversity in plants with medicinal importance around the globe and this pool of genetic variation can be considered as the base for selection, as well as for plant improvement. Xu et al. (2019) concluded that horticultural plants provide humans with various valuable products, so usage of genome editing to improve horticultural crops has substantially boosted in recent years, in different parts of the world. Hao and Xiao (2015) revealed that medicinal plants have long been utilized in traditional medicine and ethnomedicine world, and understanding of medicinal plant genome, phylogeny, and evolution are important factors to expand the knowledge pedigree, and enable the molecular breeding of medicinal plants and the sustainable utilization of plant pharmaceutical resources. Goji (Lycium barbarum) is a famous Chinese traditional super-fruit, which contains many nutrients, which has huge health benefits that attract good international markets; it is considered as both super-fruit and super-food in traditional Chinese herbal medicine and it should be part of daily diet in a sustainable life (Shahrajabian et al., 2018; Sun et al., 2019a). Ginger (Zingiber officinale) has been used as a spice and a medicine for more than 2000 years in traditional Chinese medicine (Shahrajabian et al., 2019a). In both traditional and modern Chinese medicine, ginger is used in about half of all herbal prescriptions (Shahrajabian et al., 2019b). Ginseng (Panax ginseng) is a famous herb in traditional Chinese medicine and pharmacological activities and ginseng extracts have effects on the central nervous system and largely consumed as adaptogenic herb (Shahrajabian et al., 2019c). This medicinal plant is the unique source of various types of chemical compounds, which are responsible for the various activities of the plants, and more researches on P. ginseng should be considered for the control of various diseases (Shahrajabian et al., 2019d). China has important potential to produce aromatic and medical plants and herbs due to its various biological diversity and different climatic conditions (Shahrajabian et al., 2019e,f,g; Sun et al., 2019b,c). The goal of this review is to present a short review on plant genomes of some important traditional Chinese herbs and fruits.
Ginseng
Ginseng (Panax ginseng C.A. Meyer) is a perennial herb of the Panax genus in Araliaceae family and has widely been used as a traditional medicine in Asia and other parts of the world, especially in China (Jayakodi et al., 2018). Ginseng is known to be tetraploid (2n = 4x = 48), with an estimated genome size of approximately 3.6 Gbp (Waminal et al., 2012; Choi et al., 2014).
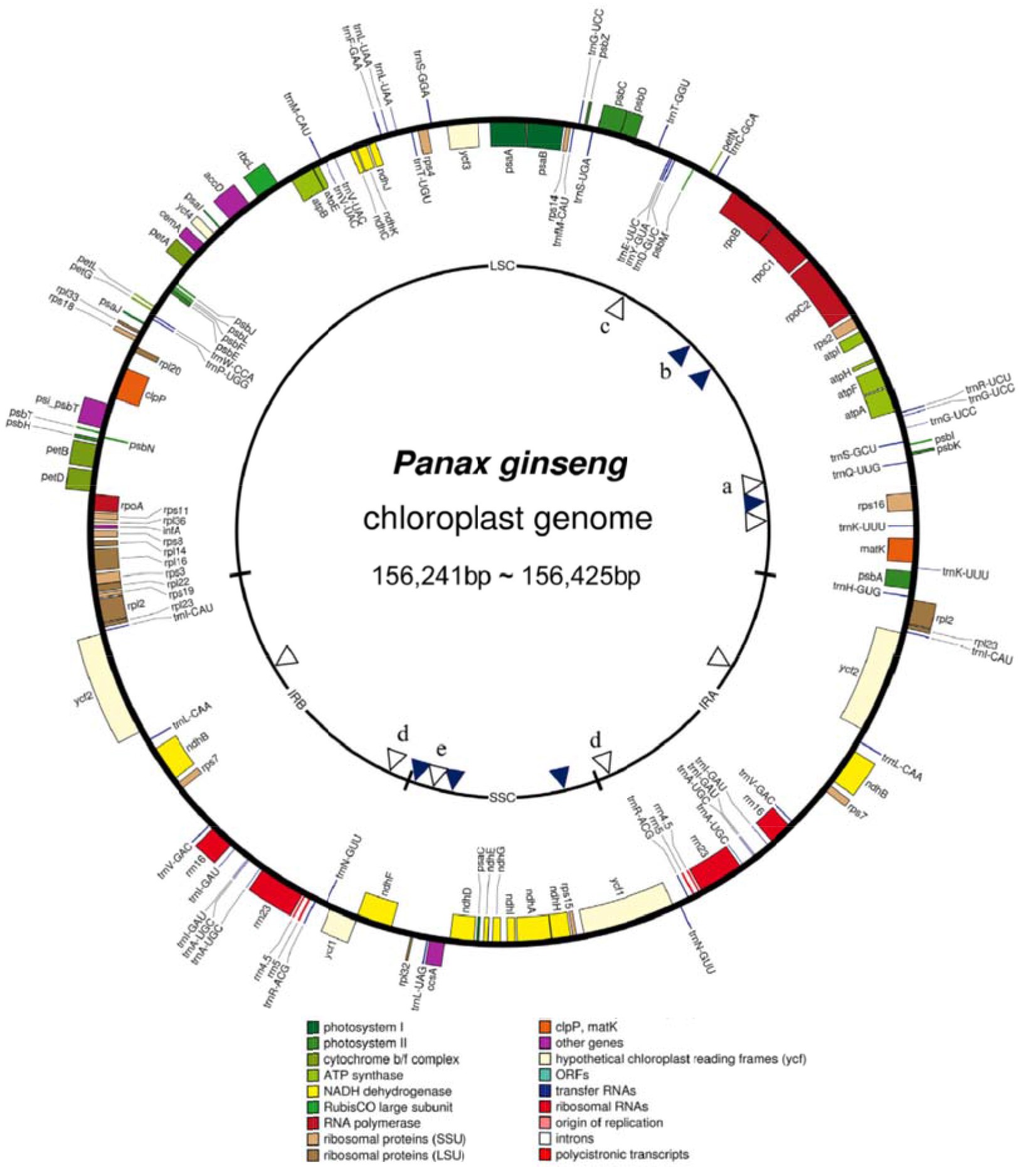
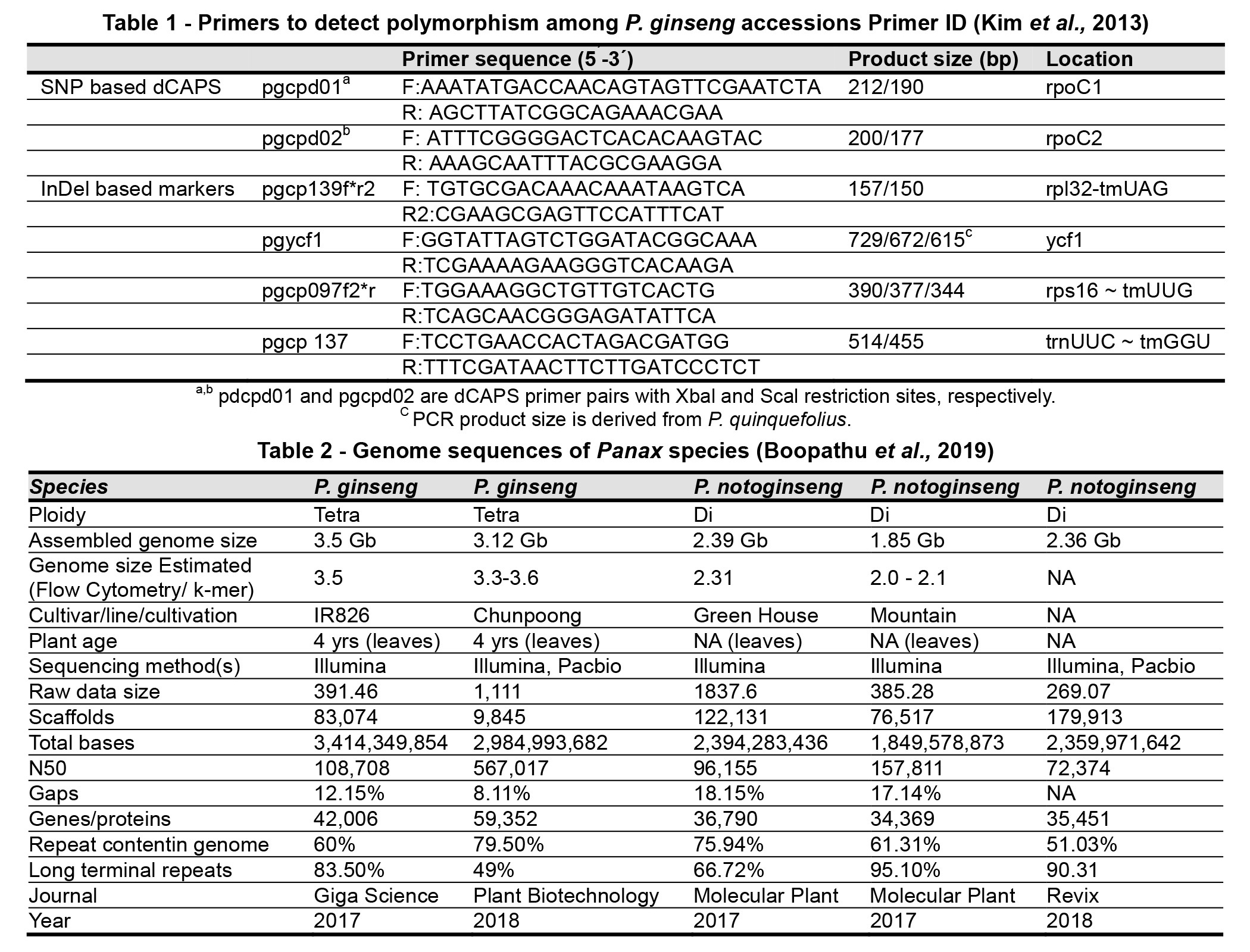
Its large, highly repetitive genome, which has experienced whole-genome duplication, has impeded the progress of whole-genome sequencing of P. ginseng (Choi et al., 2014). Chloroplast (cp) genome and 45S nuclear ribosomal DNA (45S nrDNA) sequences are the main molecular targets used for plant taxonomy, because these sequences are conserved across plant species and show clear inter-species polymorphism, whereas intra-species polymorphism is rare (Kim et al., 2013). For Panax species, they previously identified 60 polymorphic sites at the inter-species level among 101 IGS regions of three Panax species, namely P. ginseng, P. quinquefolius and P. notoginseng, using high resolution melting (HRM) analysis (Kim et al., 2015). They have obtained complete sequences of cp and 45S nrDNA, the representative barcoding target sequences for cytoplasm and nuclear genome, respectively, based on low coverage NGS sequence of each cultivar. The cp genomes sizes ranged from 156, 241 to 156, 425 bp and the major size variation was derived from differences in copy number of tandem repeats in the ycf1 gene and in the intergenic regions of rps16-trnUUG and rpl32-tmUAG. The complete 45S nrDNA unit sequences were 11,091 Bp, representing a consensus single transcriptional unit with an intergenic spacer region. Comparative analysis of these sequences, as well as those previously reported for three Chinese accessions, identified very rare, but unique polymorphism in the cp genome within P. ginseng cultivars. There were 12 intra-species polymorphisms (six SNPs and six InDels) among 14 cultivars. Chloroplast genome map of 11 P. ginseng cultivars is shown in Fig. 1. Primers of polymorphism among P. ginseng accessions Primer ID is presented in Table 1.
Obae et al. (2011) found that explants from different root morphotypes (lines) of P. quinquefolius have different callus induction response, callus growth and ginsenoside production in vitro. Also, these results show positive and significant correlations between initial total ginsenoside content of explants and callus growth and ginsenoside content of calluses, which implies that initial ginsenoside content of explants donor plants could have a significant influence on in vitro callus growth and ginsenoside production and therefore should be considered when selecting stock plants for in vitro culture. The presented results also show variability in ginsenoside profiles and genetics among P. quinquefolius lines. In spite of the fact that, no specific link between a particular DNA band or banding patterns and a given ginsenoside profile or abundance was found, this foundational research needs to be expanded to identify if there is unique DNA fingerprints can be associated with desired ginsenoside profiles in this species.
Such DNA fingerprints could be useful in selecting stock plants for use as explants for in vitro ginsenoside production or for breeding of desired cultivars of this economically important medicinal crop. Relationship tree of individual stock plants (lines) of American ginseng (Panax quinquefolius L.) revealed by unweighted pair group method with arithmetic average is presented in Fig.2.
Kim et al. (2018) indicated that diploid Panax species diverged in association with global warming in Southern Asia, and two North American species evolved via two intercontinental migrations.
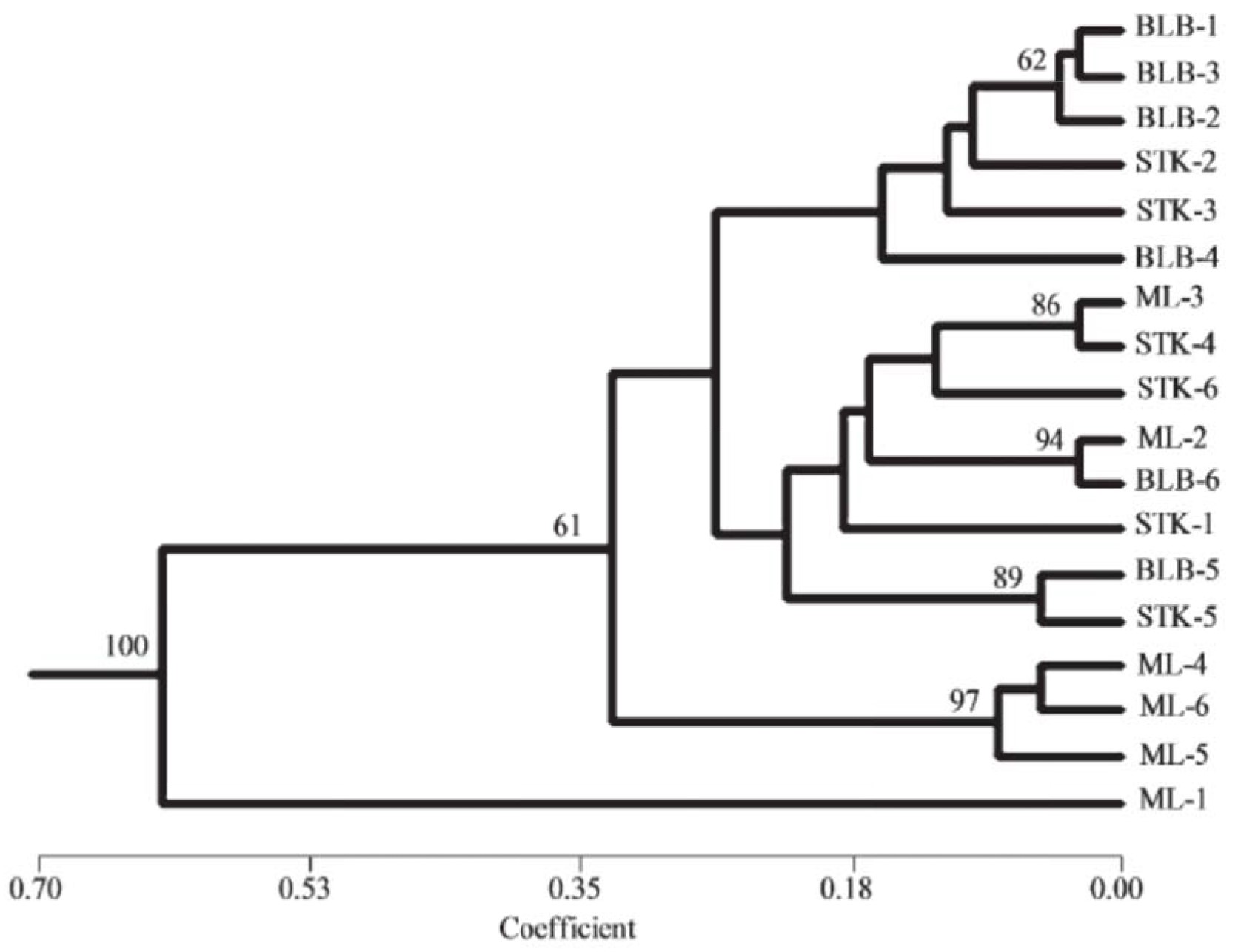
Figure 2 – Relationship tree of individual stock plants (lines) of American ginseng (Panax quinquefolius L.) revealed by unweighted pair group method with arithmetic averages (Kim et al. 2018).
Two whole genome duplications (WGD) occurred in the family Araliaceae (including Panax) after divergence with the Apiaceae, the more recent one contributing to the ability of P. ginseng to over winter, enabling it to spread broadly through the Northern Hemisphere. Functional and evolutionary analyses suggest that production of pharmacologically important dammarane-type ginsenosides originated in Panax and are produced largely in shoot tissues and transported to roots; that newly evolved P. ginseng fatty acid desaturases increase freezing tolerance, and that unprecedented retention of chlorophyll a/b binding protein genes enables efficient photosynthesis under low light. Gene map of Panax ginseng DMY chloroplast genome sequence is shown in Fig. 3.
The ginsenoside biosynthesis pathway can also be implemented in the microbial system, such as yeast to produce various type of ginsenosides in a massive quantity using the raw materials from industries as implemented for the opioids in Papaver somniferum. One of the major concerns for taking ginseng as a medicinal drug is their completely unproven myths on its toxic effects. Currently, the protein data for Panax have limitations, where those produced from a few studies are not able to be used for further studies. Hence, in future, the field of proteomics is expected to emerge with optimized high-throughput sequencing techniques to produce large-scale data sets than the earlier ones, and it has to be considered for improving the annotation of Panax genome (Boopathu et al., 2019). Genome sequences of Panax species is shown in Table 2.
Figure 3 – Gene map of Panax ginseng DMY chloroplast genome sequence. Genes shown outside the outer circle are transcribed clockwise, and those inside are transcribed counterclockwise. Genes belonging to different functional groups are color coded. The dashed area in the inner circle indicates GC content of the chloroplast (Zhao et al., 2015).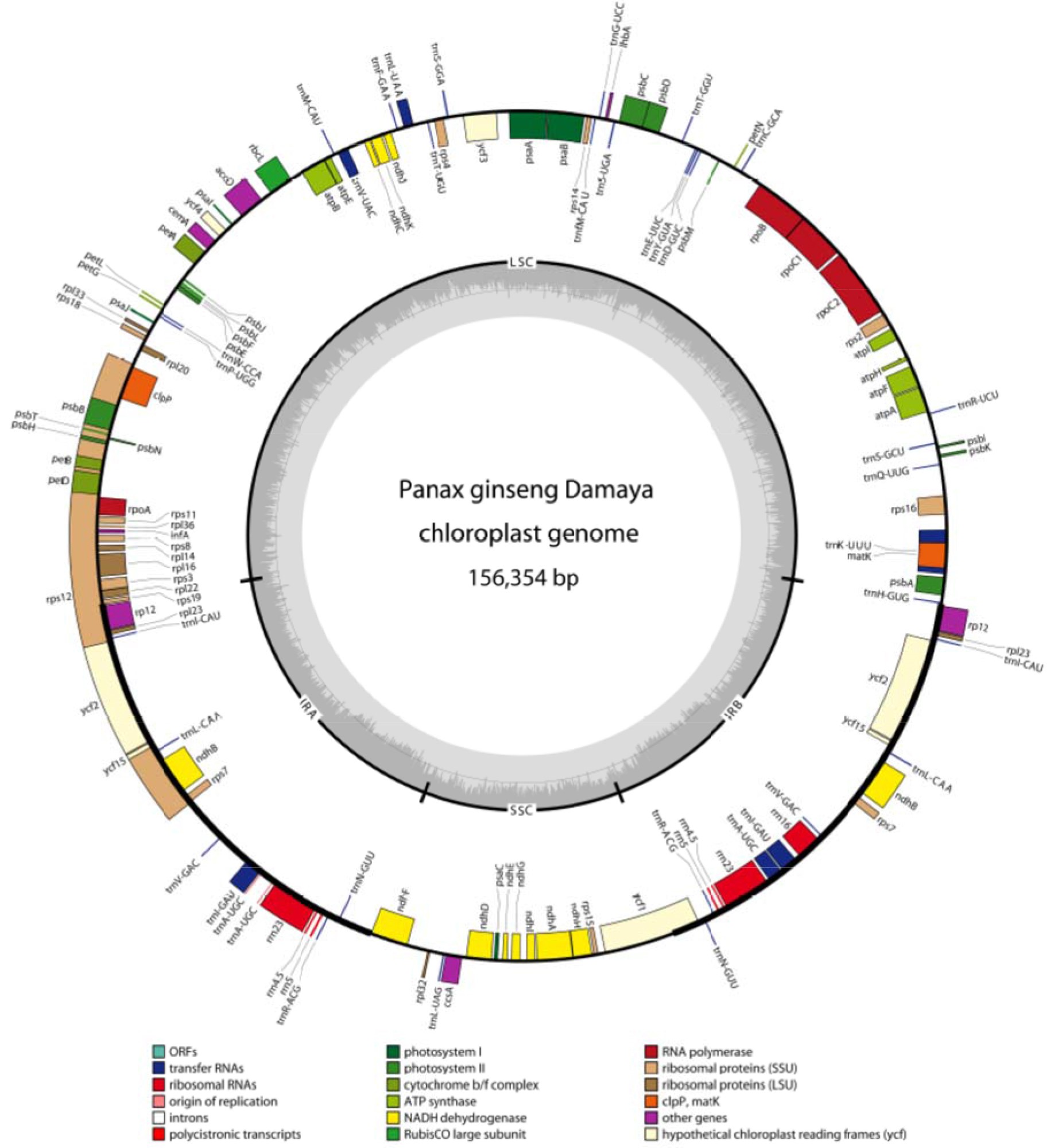
Xu et al. (2017) reported the entire genome sequence of Panax ginseng using next-generation sequencing. The 3.5-Gb nucleotide sequence contains more than 60% repeats and encodes 42006 predicted genes. A number of 22 transcriptome datasets and mass spectrometry images of ginseng roots were adopted to precisely quantify the functional genes; 31 genes were identified to be involved in the mevalonic acid pathway; eight of these genes were annotated as 3-hydroxy-3-methylgluatryl-CoA reductases, which displayed diverse structures and expression characteristics. The total 225 UDP-glycosyltransferases (UGTs) were recognized, and these UGTs accounted for one of the largest gene families of ginseng. Tandem repeats contributed to the duplication and divergence of UGTs. Molecular modeling of UGTs in the 71st, 74th, and 94th families revealed a regiospecific conserved motif located at the N-terminus. Molecular docking predicted that this motif captures ginsenoside precursors. The ginseng genome represents a valuable resource for understanding and improving the breeding, cultivation, and synthesis biology of this important herb. P. ginseng genome assembly and functional gene annotations are presented in Fig. 4.
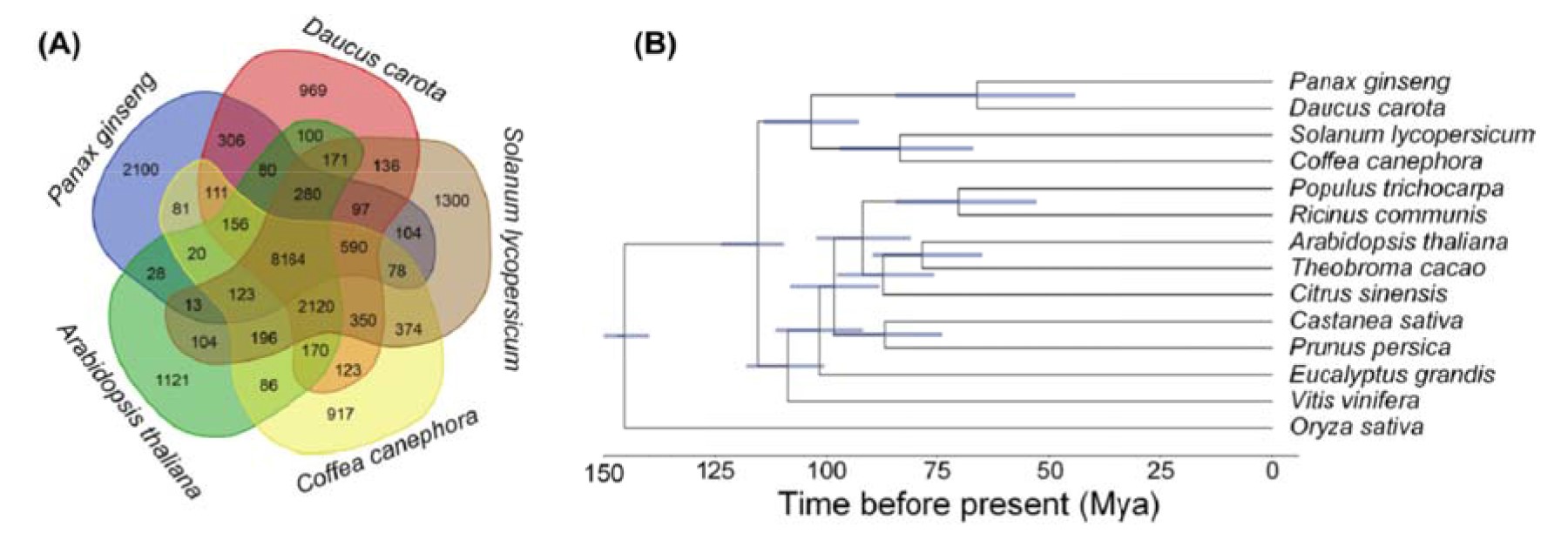
Figure 4 – P. ginseng genome assembly and functional gene annotations: (A) Phylogenetic tree and divergence data of 14 species, including P. ginseng, based on the proteins of 383 single-copy genes annotated to the genome sequence of each species; (B) Distribution of orthologous gene families in P. ginseng and four sequenced species: carrot (Daucus carota), coffee (Coffea canephora), Arabidopsis (Arabidopsis thaliana), and tomato (Solanum lycopersicum) (Xu et al., 2018).
Goji berry
Wetters et al. reported morphological and molecular based plans for the differentiation of Lycium barbarum and Lycium chinense. The two different Goji species vary significantly in seed size, with an almost double average seed area in Lycium chinense, compared to Lycium barbarum. Differences could be traced on the molecular level as well; using the psbA-trnH barcoding marker, they detected a single nucleotide substitution that was used to develop an easy one-step differentiation tool based on ARMS (amplification refractory mutation system). Two diagnostic primers used in distinct multiplex PCRs yield a second diagnostic band in a subsequent gel electrophoresis for Lycium barbarum or Lycium chinense, respectively. The ARMS approach is a strong, but simple, tool to trace either of the two different Goji species. Both the morphological and the molecular analysis showed that all of the experimented commercial goji products contained fruits of the species Goji berry (var. barbarym), leading to the assumption that consumer protection is satisfactory. Phylogenetic tree, based on psbA-tmH spacer sequences of morphological identified Lycium reference plants and commercial Goji products, is shown in Fig. 5.
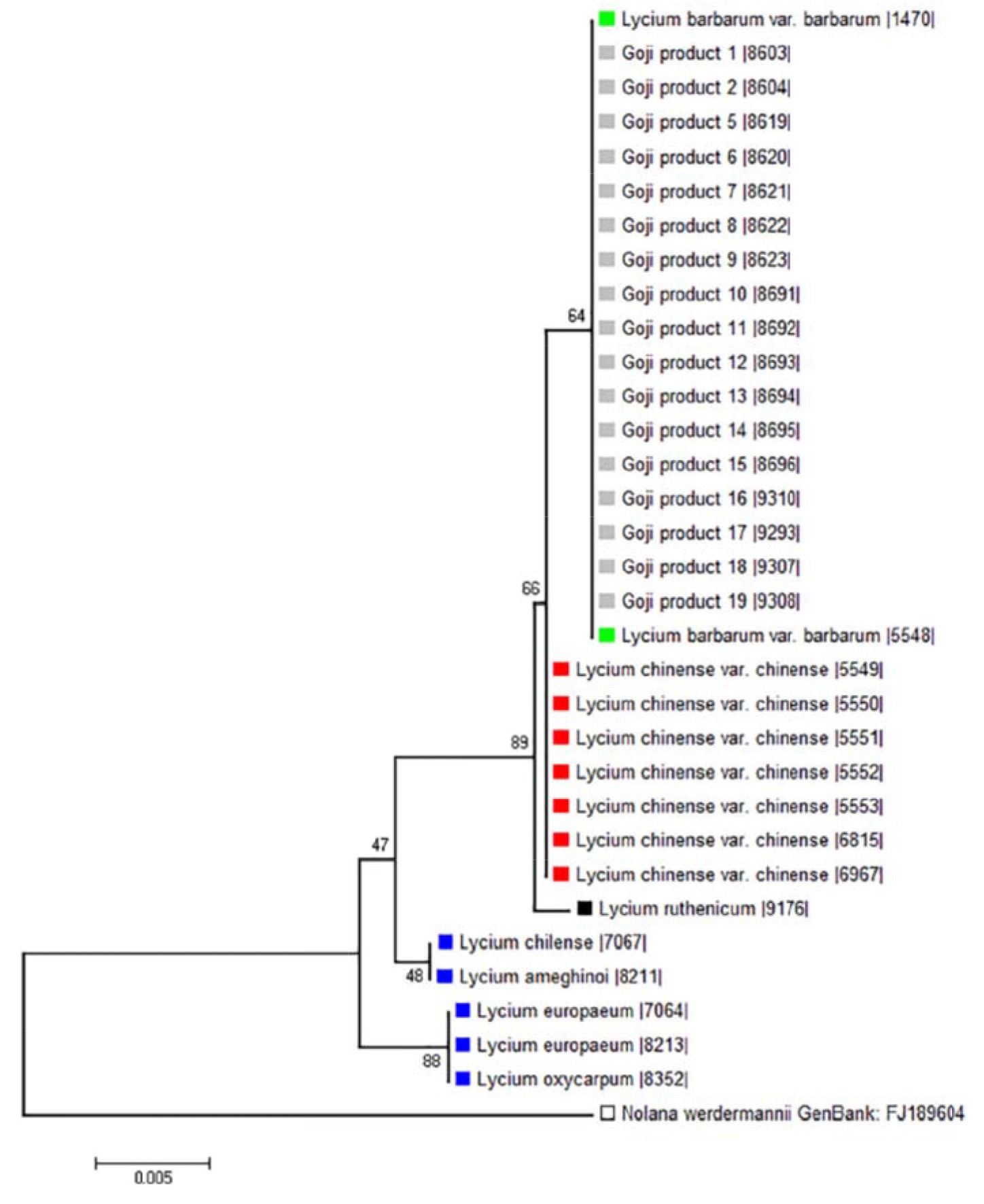
Figure 5 – Phylogenetic tree based on psbA-tmH spacer sequences of morphological identified Lycium reference plants and commercial Goji products. There were a total 532 positions in the final dataset (Wetters et al., 2018).
The fruiting stage was identified as the most responsive in the distant-grating approach and 123 miRNAs were found as up-regulating in the grafted fruit, which is remarkably higher, compared to the grafted shoot tip. Potential targets of differentially expressed miRNAs were found to be involved in diverse metabolic and regulatory pathways. ADP binding activities, molybdopterin synthase complex and RNA helicase activity were found as enriched terms of GO (Gene ontology) analysis. Additionally, metabolic pathways was revealed as the most significantly pathway in KEGG analysis. The information of the small RNA transcriptomes that are obtained might be first miRNAs elucidation for a distant-grafting system, particularly between goji and tomato.
Ginger
Mahdi et al. (2012) did experiment on three Malaysian ginger cultivars (Bukit Tinggi, TanjungSepat and Sabah) were collected and examined for genetic polymorphisms using microsatellite DNA primers. The single microsatellite oligonucleotide primers (CATA)5, (GATA)5 and (GAC)6 were used in polymerase chain reactions (PCRs). The PCR reactions produced seven polymorphic bands with an average of 2.334 polymorphic bands per primer, leading to an average polymorphism rate of 17.9%. Cluster analysis revealed 87.50% similarity between Bukit Tinggi and Tanjung Sepat, 64.27% similarity between Bukit Tinggi and Sabah and 56.25% similarity between Tanjung, 64.27% similarity between Bukit Tinggi. DNA sequencing of the polymorphic PCR products of Tanjung Sepat ginger revealed the characteristics features of a putative new gene: a core promoter sequence, an enhancer and a transcription start site. Cluster analysis using the unweighted pair group method with arithmetic average (UPGMA) was used to construct a phylogenetic tree, which indicated that Bukrit Tinggi ginger is genetically more closely related to Tanjung Sepat ginger than to Sabah ginger. On the basis of their results, they conclude that there is genotypic variation among ginger cultivars, and the microsatellite DNA polymerase useful for detecting polymorphic DNA in Malaysian ginger cultivars. Prasath et al. (2014) found that a total of 36359 and 32312 assembled transcript sequences were obtained from both Curcuma amada and ginger. The roles of the unigenes cover a diverse set of molecular functions and biological processes, among which they identified a large number of genes associated with resistance to stresses and response to biotic stimuli. Large-scale expression profiling showed that many of the disease resistance related genes were expressed more in C. amada. Comparative analysis also identified genes belonging to different pathways of plant defense against biotic stresses that are differentially expressed in either ginger or mango ginger. The identification of many defense related genes differentially expressed provides many insights to the resistance mechanism to Ralstonia solanacearum and for studying potential pathways involved in responses to pathogens. Also, several candidate genes that may highlight the difference in resistance to R. solanacearum between ginger and mango ginger were identified. Divergence in the expression levels of transcription factors (TF) transcripts between C. amada and Zingiber officinale is shown in Fig. 6.
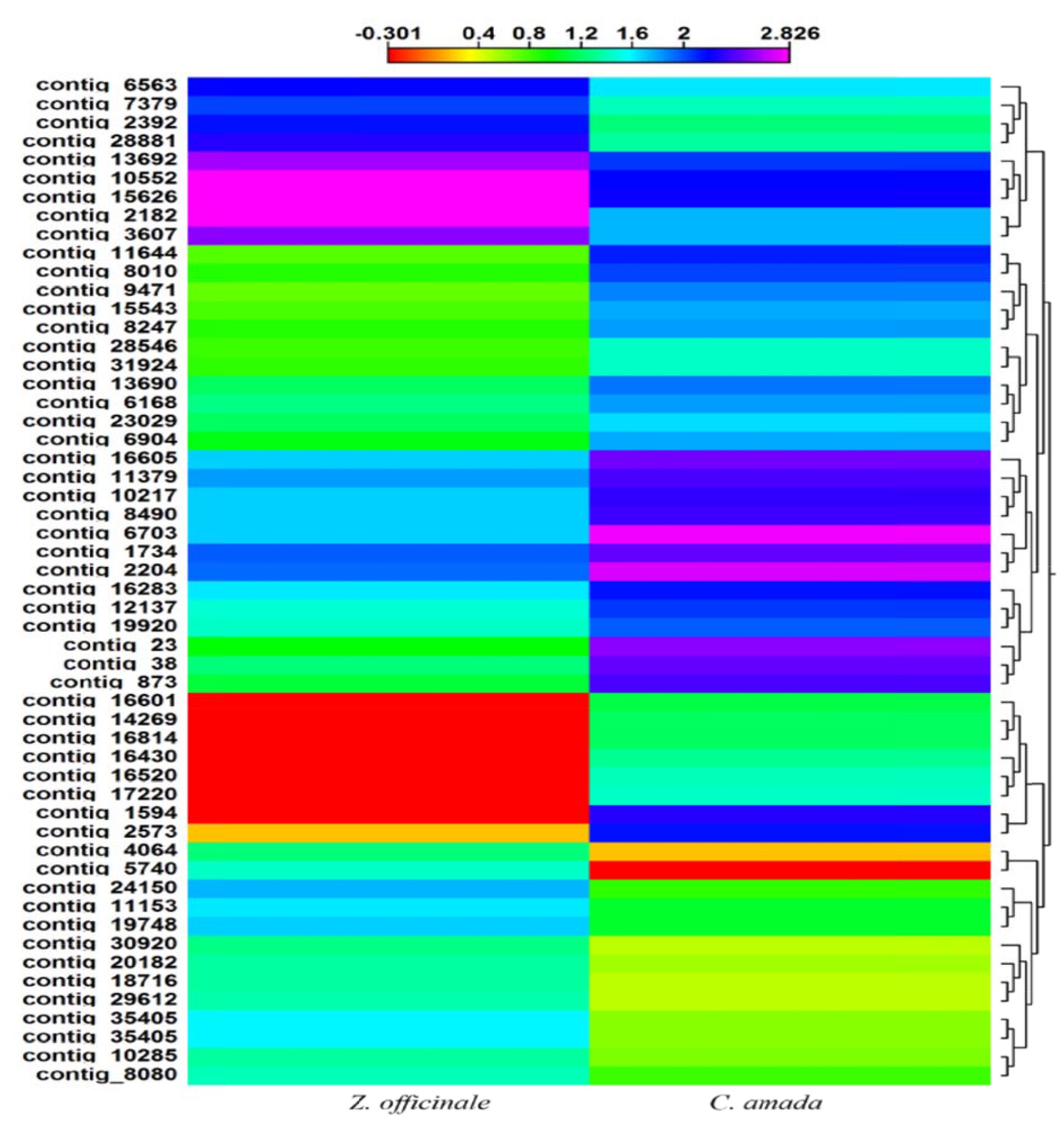
Figure 6 – Divergence in the expression levels of transcription factors (TF) transcripts between C. amada and Z. officinale (Prasath et al., 2014)
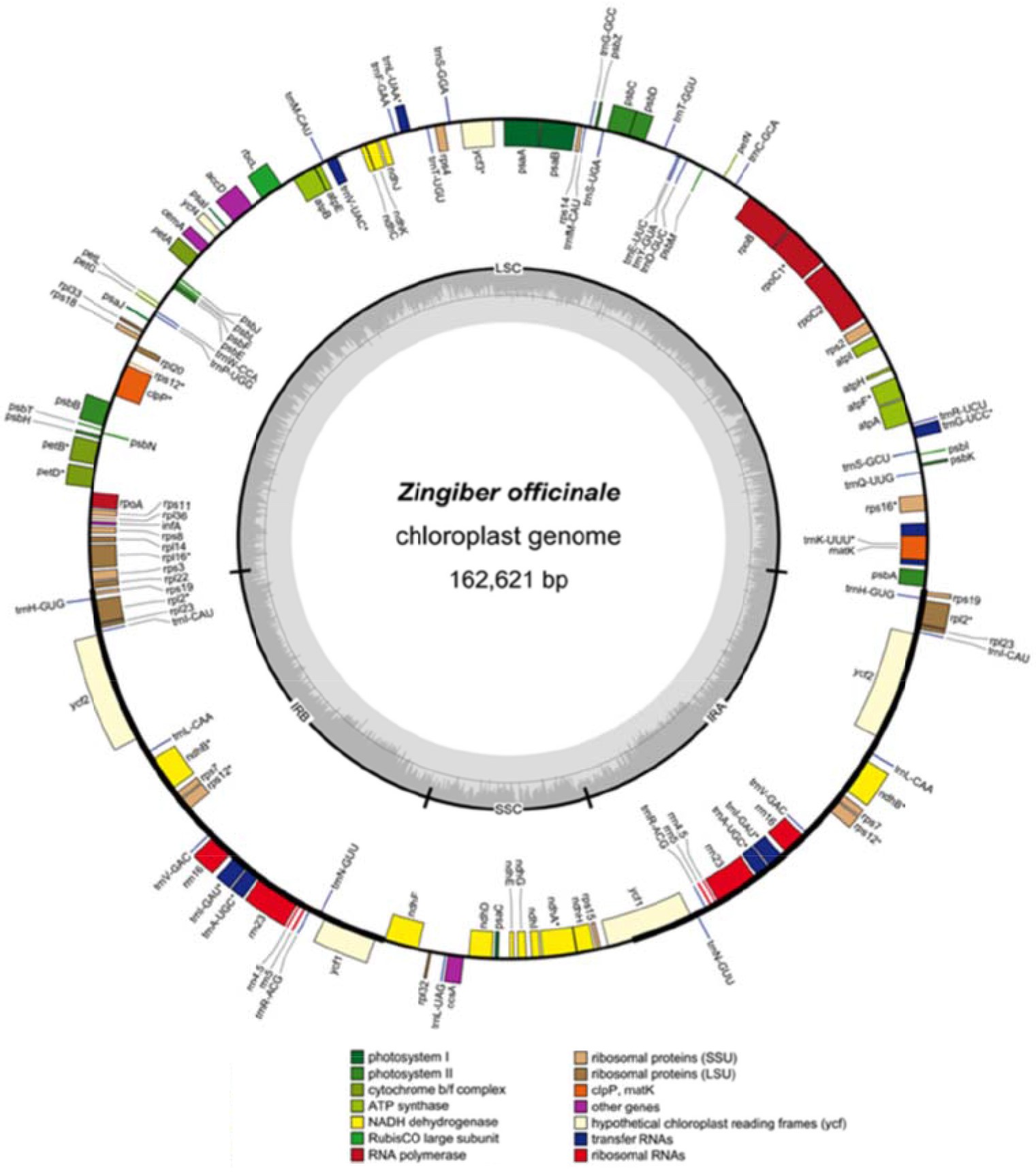
Cui et al. (2019) sequenced and analyzed the Z. officinale chloroplast genome. Firstly, the basic structures, gene information, and codon usage pattern were revealed. Secondly, 78 SSRs and 42 long repeat sequences were identified. Thirdly, a comparative analysis within the family Zingiberaceae was executed and some variable regions, which have the potential to become DNA markers were revealed. The results showed that small single-copy (SSC) was the most variable region and may be undergoing rapid nucleotide substitution in Zingiberaceae species. The ML tree indicated that the Zingiber species has a close relationship with species in Kaempferia and Curcuma and clearly show a phylogenetic relationship with species in the family Zingiberaceae, or even the order Zingiberaceae, through molecular methods. Gene map of the Zingiber officinale complete chloroplast genome is shown in Fig. 7.
CONCLUSIONS
Traditional Chinese medicine (TCM), is an essential part of the health care system in many countries in sustainable way, which relies on natural products and has been playing a very important role in health protection and disease control for many years. Few studies have been carried out on genomics for medicinal plants. Plant genomes are characterized by large variations of genome size and ploidy level. Comparative genomics provides a method to unravel the relationship between genomes by describing conserved chromosomes or chromosomal regions between related species. There has been a significant increase is the number of sequenced genomes of medicinal plant species. It is also clear that it is possible to use plant genome as a tool for improving breeding strategies. More researches are needed to introduce the genome research of medicinal plants, including genome sequencing, assembly, annotation, and functional genomics to set up the foundation for progress and development of natural medicines and also the appropriate selection of cultivars with good agricultural traits.
REFERENCES
Boopathu, V., Subramaniyam, S., Mathiyalagan, R. & Yang, D.-C. (2019). Till 2018: a survey of biomolecular sequences in genus Panax. J Ginseng Res., 44(1): 33-43, DOI: 10.1016/j.jgr.2019.06.004
Choi, H.I., Waminal, N.E., Park, H.M., Kim, N.H., Choi, B.S., Park, M., Choi, D., Lim, Y.P., Kwon, S.J. et al. (2014). Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy. Plant J., 77(6): 906-916, DOI: 10.1111/tpj.12441.
Cui, Y., Nie, L., Sun, W., Xu, Z., Wang, Y., Yu, J., Song, J. & Yao, H. (2019). Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family Zingiberaceae based on the complete chloroplast genome. Plants (Basel), 8(8): 283, DOI: 10.3390/plants8080283
Gantait, S., Debnath, S. & Ali M.N. (2014). Genomic profile of the plants with pharmaceutical value. 3 Biotech, 4: 563-578, DOI: 10.1007/ s13205-014-0218-9
Hao, D.-C. & Xiao, P.-G. (2015). Genomics and evolution in traditional medicinal plants: road to a healthier life. Evol.Bioinf., 11:197-212, DOI: 10.4137/EBO.S31326
Jayakodi, M., Choi, B.-S., Lee, S.-C., Kim, N.-H., Park, J.Y., Jang, W., Lakshmanan, M., Mohan, S.V.G., Lee, D.-Y. & Yang, T.-J. (2018). Ginseng genome database: an open-access platform for genomics of Panax ginseng. BMC Plant Biol., 18(1): 62, DOI: 10.1186/s12870-018-1282-9
Jiang, F., Zhang, J., Wang, S., Yang, L., Luo, Y., Gao, S., Zhang, M., Wu, S., Hu, S., Sun, H. & Wang, Y. (2019). The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic.Res., 6(1): 128, DOI: 10.1038/s41438-019-0215-6
Kim, J.H., Jung, J.-Y., Choi, H.-I., Kim, N.-H, Park, J.Y., Lee, Y. et al. (2013). Diversity and evolution of major Panax species revealed by scanning the entire chloroplast intergenic spacer sequences. Genet.Resour.Crop Evol., 60: 413-425. DOI: 10.1007/s10722-012-9844-4
Kim, K., Lee, S.-C., Lee, J., Lee, H.-O, Joh, H.J., Kim, N.-H.. et al. (2015). Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One, 10(6): e0117159, DOI: 10.1371/journal. pone.0117159
Kim, N.-H., Jayakodi, M., Lee, S.-C., Choi, B.-S., Jang, W., Lee, J., Kim, H.H. et al. (2018). Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J., 16(11): 1904-1917, DOI: 10.1111/pbi.12926
Mahdi, H.J., Andayani, R. & Aziz, I. (2013). Determination of phylogenetic and molecular characteristics of three Malaysian ginger cultivars (Zingiber officinale Roscoe) using microsatellite DNA. Trop Life Sci., 24(2): 65-76.
Obae, S.G., Klandorf, H. & West, T.P. (2011). Growth characteristics and ginsenosides production of in vitro tissues of American ginseng, Panax quinquefolius L. HortSci., 46(8): 1136-1140, DOI: 10.21273/HORTSC I.46.8.1136
Ogbaji, P.O., Shahrajabian, M.H. & Xue, X. (2013). Changes in germination and primarily growth of three cultivars of tomato under diatomite and soil materials in auto-irrigation system. Int.J.Biol., 5(3):80-84, DOI: 10.5539/ijb.v5n3p80
Ogbaji, P.O., Li, J., Xue, X., Shahrajabian, M.H. & Egrinya, E.A. (2018). Impact of bio-fertilizer or nutrient on spinach (Spinaceae oleracea) growth and yield in some province soils of P.R. China. Cercet.Agron. in Moldova, 51(2): 43-52, DOI: 10.2478/cerce-2018-0015
Prasath, D., Karthika, R., Habeeba, N.T., Suraby, E.J., Rosana, O.B., Shaji, A., Eapen, S.J., Deshpande, U., Anandaraj, M. (2014). Comparison of the transcriptomes of ginger (Zingiber officinale Rosc.) and mango ginger (Curcuma amada Roxb.) in response to the bacterial wilt infection. PLoS One, 9(6): e99731, DOI: 10.1371/journal.pone. 0099731
Shahrajabian, M.H., Sun, W. & Cheng, Q.A. (2018). A review of goji berry (Lycium barbarum) in traditional Chinese medicine as a promising organic superfood and superfruit in modern industry. Acad.J.Med. Plants, 6(12): 437-445, DOI: 10.15 413/ajmp.2018.0186
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2019a). Pharmacological uses and health benefits of ginger (Zingiber officinale) in traditional Asian and ancient Chinese medicine, and modern practice. Not.Sci.Biol., 11(3): 309-319, DOI: 10.15835/nsb11310419
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2019b). Clinical aspects and health benefits of ginger (Zingiber officinale) in both traditional Chinese medicine and modern industry. Acta Agr.Scand.B-S.P., 69(6): 546-556, DOI: 10.1080/09064710.2019.1606 930
Shahrabian, M.H., Sun, W. & Cheng, Q. (2019c). A review of ginseng species in different regions as a multipurpose herb in traditional Chinese medicine, modern herbology and pharmacological science. J.Med.Plant Res., 13(10): 213-226, DOI: 10.5897/JMPR2019. 6731
Shahrajabian, M.H., Sun, W., Cheng, Q. (2019d). The power of natural Chinese medicine, ginger and ginseng root in an organic life. Middle East J.Sci.Res., 27(1): 64-71, DOI: 10.5829/idosi.mejsr.2019.64.71
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2019e). A review of astragalus species as foodstuffs, dietary supplements, a traditional Chinese medicine and a part of modern pharmaceutical science. Appl.Ecol. Env.Res., 17(6): 13371-13382, DOI: 10.15666/aeer/1706_1337113382
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2019f). Sustainable agriculture and soybean, a legume in traditional Chinese with great biological nitrogen fixation. J.Biol.Environ.Sci., 13(38): 71-78.
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2019g). Tremendous health benefits and clinical aspects of Smilax china. Afr. J.Pharm. Pharmacol., 13(16): 253-258, DOI: 10.5897/AJPP2019.5070
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2020a). Chinese star anise (Illicium verum) and pyrethrum (Chrysanthemum cinerariifolium) as natural alternatives for organic farming and health care – a review. Austr.J.Crop Sci., 14(03): 517-523, DOI: 10.21475/ajcs.20.14.03.p2209
Shahrajabian, M.H., Sun, W., Shen, H. & Cheng, Q. (2020b). Chinese herbal medicine for SARS and SARS-CoV-2 treatment and prevention, encouraging using herbal medicine for COVID-19 outbreak. Acta Agr.Scand.B-S.P., 70(5): 437-443, DOI: 10.1080/09064710.2020.1763 448
Shahrajabian, M.H., Sun, W. & Cheng, Q. (2020c). A short review of goji berry, ginger, ginseng and astragalus in traditional Chinese and Asian medicine. BSJ Health Sci., 3(2).
Soleymani, A. & Shahrajabian, M.H. (2012). Response of different cultivars of fennel (Foeniculum vulgare) to irrigation and planting dates in Isfahan, Iran. Res. Crops, 13(2): 656-600.
Soleymani, A. & Shahrajabian, M.H. (2018). Changes in germination and seedling growth of different cultivars of cumin to drought stress. Cercet.Agron. in Moldova, 51(1): 91-100, DOI: 10.2478/cerce-2018-0008
Sun, W., Shahrajabian, M.H. & Cheng, Q. (2019a). Therapeutic roles of goji berry and ginseng in traditional Chinese. J.Nutr. Food Secur., 4(4): 293-305, DOI: 10.18502/jnfs.v4i4. 1727
Sun, W., Shahrajabian, M.H. & Cheng, Q. (2019b). The insight and survey on medicinal properties and nutritive components of shallot. J.Med. Plant Res., 13(18): 452-457, DOI: 10.58 97/JMPR2019.6836
Sun, W., Shahrajabian, M.H. & Cheng, Q. (2019c). Anise (Pimpinella anisum l.), a dominant spice and traditional medicinal herb for both food and medicinal purposes. Cogent Biol., 5(1): 1-25, DOI: 10.1080/23312025.2019.1673688
Sun, W., Shahrajabian, M.H., Khoshkharam, M. & Cheng, Q. (2020a). Adaptation of acupuncture and traditional Chinese herbal medicines models because of climate change. J. Stress Physiol. Biochem., 16(1): 85-90.
Sun, W., Shahrajabian, M.H., Khoshkharam, M., Shen, H. & Cheng, Q. (2020b). Cultivation of cotton in China and Iran with considering biological activities and its health benefits. Cercet.Agron. in Moldova, 53(1):105-120, DOI: 10.2478/cerce-2020-0005
Waminal, N.E, Park, H.M., Ryu, K.B., Kim, J.H., Yang, T.-J. & Kim, H.H. (2012). Karyotype analysis of Panax ginseng C.A. Meyer, 1843 (Araliaceae) based on rDNA loci and DAPI and distribution. Comp. Cytogenet., 6(4): 425-441, DOI: 10.3897/CompCytogen.v6i4.3740
Wetters, S., Horn, T. & Nick, P. (2018). Goji who? Morphological and DNA based authentication of a superfood. Front. Plant Sci., 9:1859, DOI: 10.3389/fpls.2018.01859
Xiong, C., Sun, W., Li, J., Yao, H., Shi, Y., Wang, P., Huang, B., Shi, L., Liu, D., Hu, Z. & Chen, S. (2018). Identifying the species of seeds in traditional Chinese medicine using DNA barcoding. Front Pharmacol., 9: 701, DOI: 10.3389/fphar.2018.00701
Xu, J., Chu, Y., Liao, B., Xiao, S., Yin, Q., Bai, R., Su, H., Dong, L. et al. (2017). Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience, 6(11): 1-15, DOI: 10.1093/gigascience/gix 093
Xu, J., Hua, K. & Lang, Z. (2019). Genome editing for horticultural crop improvement. Hortic.Res., 6: 113, DOI: 10.1038/s41438-019-0196-5
Zhao, Y., Yin, J., Guo, H., Zhang, Y., Xiao, W., Sun, C., Wu, J., Qu, X., Yu, J., Wang, X. & Xiao, J. (2015). The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci., 5(1): 696, DOI: 10.3389/fpls.2014.00696